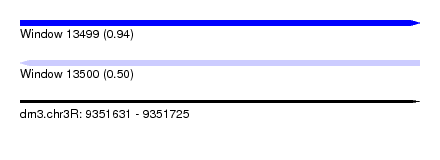
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,351,631 – 9,351,725 |
| Length | 94 |
| Max. P | 0.939997 |

| Location | 9,351,631 – 9,351,725 |
|---|---|
| Length | 94 |
| Sequences | 12 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 69.13 |
| Shannon entropy | 0.63732 |
| G+C content | 0.48565 |
| Mean single sequence MFE | -28.12 |
| Consensus MFE | -9.43 |
| Energy contribution | -9.52 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.939997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9351631 94 + 27905053 -------------UGCACCAACCGCCGACCA-UUCCAAGUGGCCAGU-CGCACGUUAUAUCGUUAAUGAAAACUGAUUCUAGUGUCAUUAACGUGCGAGCUCUCCAGAC -------------.((.......(((.((..-......)))))...(-((((((......)((((((((..((((....)))).))))))))))))))))......... ( -24.00, z-score = -1.91, R) >droSim1.chr3R_random 536639 94 + 1307089 -------------UGCACCAACCGCCGACCA-UUCCGAGUGGCCAGU-CGCACGUUAUAUCGUUAAUGAAAACUGAUUCUAGUGUCAUUAACGUGCGAGCUCUCCAGAC -------------.((.....((((((....-...)).))))....(-((((((......)((((((((..((((....)))).))))))))))))))))......... ( -25.90, z-score = -2.11, R) >droSec1.super_0 8465218 94 - 21120651 -------------UACACCAACCGCCGACCA-UUCCGAGUGGCCAGU-CGCACGUUAUAUCGUUAAUGAAAACUGAUUCUAGUGUCAUUAACGUGCGAGCUCUCCAGAC -------------........((((((....-...)).))))....(-((((((......)((((((((..((((....)))).))))))))))))))........... ( -24.00, z-score = -1.92, R) >droEre2.scaffold_4770 5455242 94 - 17746568 -------------AGCACCAACUGCCGACCA-UUCCGAGUGGGCAGU-CGCACGUUAUAUCGUUAAUGAAAACUGAUUCUAGUGUCAUUAACGUGCGAGCUCUCCAGAC -------------(((.....(((((.((..-......)).)))))(-((((((......)((((((((..((((....)))).)))))))))))))))))........ ( -31.40, z-score = -3.30, R) >droYak2.chr3R 13658672 94 + 28832112 -------------UGCAGCAACCGCCGACCA-UUCCGAGUGGGCAGU-CGCACGUUAUAUCGUUAAUGAAAACUGAUUCUAGUGUCAUUAACGUGCGAGCUCUCCAGAC -------------...(((....(((.((..-......)).)))..(-((((((......)((((((((..((((....)))).)))))))))))))))))........ ( -28.40, z-score = -1.99, R) >droAna3.scaffold_13340 1905113 87 + 23697760 ---------------------CAGCUCACCACUUCCGAGUGGCCAGU-GGCACGUUAUAUCGUUAAUGAAAACUGAUUCUAGUGUCAUUAACGUGCGAGCUUUUCAGAC ---------------------.(((((.(((((....))))).....-.(((((......)((((((((..((((....)))).)))))))))))))))))........ ( -27.60, z-score = -2.98, R) >dp4.chr2 16793304 109 + 30794189 CGCACAAAGUGGUGCUCUCGACGAGUCCGCGAACGCGUCCGAGUGCCCGCCACGUUAUAUCGUUAAUGAAAACUGAUUCUAGUGUCAUUAACGUGCGAGCUUUUCAGAC (((((...(((((((.(((((((.((......)).))).)))).))..)))))........((((((((..((((....)))).)))))))))))))............ ( -38.10, z-score = -2.77, R) >droPer1.super_0 8222326 109 - 11822988 CGCACAAAGUGGUGCUCUCGACGAGUCCGCGAACGCGUCCGAGUGCCCGCCACGUUAUAUCGUUAAUGAAAACUGAUUCUAGUGUCAUUAACGUGCGAGCUUUUCAGAC (((((...(((((((.(((((((.((......)).))).)))).))..)))))........((((((((..((((....)))).)))))))))))))............ ( -38.10, z-score = -2.77, R) >droWil1.scaffold_181130 6420650 98 + 16660200 -------AGCACUUCACUUGGCGAA---ACGGGUCUGAU-GACUGCCCGGUAGGGUAUAUCGUUAAUGAAAACUGAUUCUAGUGUCAUUAACGUGAGAGCUUUUCAGAC -------(((.((.(((.....((.---...((((....-))))((((....))))...))((((((((..((((....)))).))))))))))))).)))........ ( -26.70, z-score = -1.10, R) >droVir3.scaffold_13047 9143485 75 + 19223366 ---------------------------------CAAGUCAGCCCACU-AGCAGAGCAUAUCGUUAAUGAAAACUGAUUCUAGUGCCAUUAACGUACGAGCUCUUCAGAC ---------------------------------...(((.((.....-.))(((((....((((((((...((((....))))..)))))))).....)))))...))) ( -19.90, z-score = -2.91, R) >droMoj3.scaffold_6540 2647900 87 - 34148556 ---------------------UUUGUGUUCGCGCAGCUCUGGCCACU-GGCGCUGCAUUUCGUUAAUGAAAACUGAUUCUAGUGCCAUUAAGGUGCGAGCUCUUCAGGU ---------------------...(.(((((((((((.(..(...).-.).)))))....(.((((((...((((....))))..)))))).).)))))).)....... ( -27.70, z-score = -1.01, R) >droGri2.scaffold_14906 10069403 92 - 14172833 ----------------CUUAACAAGAGCGAGUCCAAGUCCGGCCGCG-AGCGCGGAAUAUCGUUAAUGAAAACUGAUUCUAGUGCCAUUAACGUACGAGCUCUUCAGAC ----------------......((((((..((.(...((((((....-.)).)))).....(((((((...((((....))))..)))))))).))..))))))..... ( -25.70, z-score = -1.47, R) >consensus _____________U_CACCAACCGCCGACCA_UUCCGAGUGGCCACU_CGCACGUUAUAUCGUUAAUGAAAACUGAUUCUAGUGUCAUUAACGUGCGAGCUCUUCAGAC ............................................................((((((((...((((....))))..))))))))................ ( -9.43 = -9.52 + 0.08)
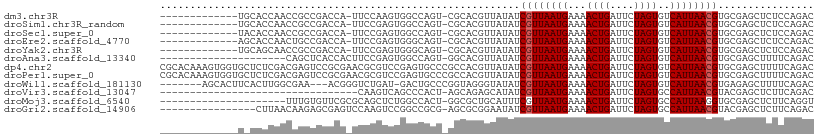

| Location | 9,351,631 – 9,351,725 |
|---|---|
| Length | 94 |
| Sequences | 12 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 69.13 |
| Shannon entropy | 0.63732 |
| G+C content | 0.48565 |
| Mean single sequence MFE | -27.68 |
| Consensus MFE | -9.90 |
| Energy contribution | -9.79 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr3R 9351631 94 - 27905053 GUCUGGAGAGCUCGCACGUUAAUGACACUAGAAUCAGUUUUCAUUAACGAUAUAACGUGCG-ACUGGCCACUUGGAA-UGGUCGGCGGUUGGUGCA------------- .............((((((((((((.(((......)))..)))))))).......(((.((-(((..((....))..-.))))))))....)))).------------- ( -26.30, z-score = -0.47, R) >droSim1.chr3R_random 536639 94 - 1307089 GUCUGGAGAGCUCGCACGUUAAUGACACUAGAAUCAGUUUUCAUUAACGAUAUAACGUGCG-ACUGGCCACUCGGAA-UGGUCGGCGGUUGGUGCA------------- .........((.(((((((((((((.(((......)))..))))))))(......))))))-((..(((.(.(((..-...)))).)))..)))).------------- ( -27.30, z-score = -0.61, R) >droSec1.super_0 8465218 94 + 21120651 GUCUGGAGAGCUCGCACGUUAAUGACACUAGAAUCAGUUUUCAUUAACGAUAUAACGUGCG-ACUGGCCACUCGGAA-UGGUCGGCGGUUGGUGUA------------- ............(((((((((((((.(((......)))..))))))))(......))))))-((..(((.(.(((..-...)))).)))..))...------------- ( -25.40, z-score = -0.13, R) >droEre2.scaffold_4770 5455242 94 + 17746568 GUCUGGAGAGCUCGCACGUUAAUGACACUAGAAUCAGUUUUCAUUAACGAUAUAACGUGCG-ACUGCCCACUCGGAA-UGGUCGGCAGUUGGUGCU------------- .............((((((((((((.(((......)))..))))))))...........((-((((((.(((.....-.))).)))))))))))).------------- ( -29.80, z-score = -1.68, R) >droYak2.chr3R 13658672 94 - 28832112 GUCUGGAGAGCUCGCACGUUAAUGACACUAGAAUCAGUUUUCAUUAACGAUAUAACGUGCG-ACUGCCCACUCGGAA-UGGUCGGCGGUUGCUGCA------------- .............((((((((((((.(((......)))..))))))))).........(((-((((((.(((.....-.))).)))))))))))).------------- ( -30.90, z-score = -1.86, R) >droAna3.scaffold_13340 1905113 87 - 23697760 GUCUGAAAAGCUCGCACGUUAAUGACACUAGAAUCAGUUUUCAUUAACGAUAUAACGUGCC-ACUGGCCACUCGGAAGUGGUGAGCUG--------------------- ........(((((...(((((((((.(((......)))..))))))))).........(((-(((..((....)).))))))))))).--------------------- ( -28.60, z-score = -3.16, R) >dp4.chr2 16793304 109 - 30794189 GUCUGAAAAGCUCGCACGUUAAUGACACUAGAAUCAGUUUUCAUUAACGAUAUAACGUGGCGGGCACUCGGACGCGUUCGCGGACUCGUCGAGAGCACCACUUUGUGCG (.((....))).(((((((((((((.(((......)))..))))))))........((((...((.((((.(((.(((....))).))))))).)).))))...))))) ( -36.10, z-score = -1.82, R) >droPer1.super_0 8222326 109 + 11822988 GUCUGAAAAGCUCGCACGUUAAUGACACUAGAAUCAGUUUUCAUUAACGAUAUAACGUGGCGGGCACUCGGACGCGUUCGCGGACUCGUCGAGAGCACCACUUUGUGCG (.((....))).(((((((((((((.(((......)))..))))))))........((((...((.((((.(((.(((....))).))))))).)).))))...))))) ( -36.10, z-score = -1.82, R) >droWil1.scaffold_181130 6420650 98 - 16660200 GUCUGAAAAGCUCUCACGUUAAUGACACUAGAAUCAGUUUUCAUUAACGAUAUACCCUACCGGGCAGUC-AUCAGACCCGU---UUCGCCAAGUGAAGUGCU------- ((((((...(((....(((((((((.(((......)))..))))))))).....(((....))).))).-.)))))).(((---(((((...))))))))..------- ( -24.80, z-score = -2.28, R) >droVir3.scaffold_13047 9143485 75 - 19223366 GUCUGAAGAGCUCGUACGUUAAUGGCACUAGAAUCAGUUUUCAUUAACGAUAUGCUCUGCU-AGUGGGCUGACUUG--------------------------------- (((...(((((..((.(((((((((.(((......)))..)))))))))))..)))))(((-....))).)))...--------------------------------- ( -21.30, z-score = -1.65, R) >droMoj3.scaffold_6540 2647900 87 + 34148556 ACCUGAAGAGCUCGCACCUUAAUGGCACUAGAAUCAGUUUUCAUUAACGAAAUGCAGCGCC-AGUGGCCAGAGCUGCGCGAACACAAA--------------------- .........(.((((.(.(((((((.(((......)))..))))))).)....((((((((-...)))....))))))))).).....--------------------- ( -20.30, z-score = 0.07, R) >droGri2.scaffold_14906 10069403 92 + 14172833 GUCUGAAGAGCUCGUACGUUAAUGGCACUAGAAUCAGUUUUCAUUAACGAUAUUCCGCGCU-CGCGGCCGGACUUGGACUCGCUCUUGUUAAG---------------- .....((((((.....(((((((((.(((......)))..)))))))))....((((.((.-....)))))).........))))))......---------------- ( -25.30, z-score = -0.80, R) >consensus GUCUGAAGAGCUCGCACGUUAAUGACACUAGAAUCAGUUUUCAUUAACGAUAUAACGUGCC_ACUGGCCACUCGGAA_UGGUCGGCGGUUGAUG_A_____________ ................(((((((((.(((......)))..)))))))))............................................................ ( -9.90 = -9.79 + -0.10)
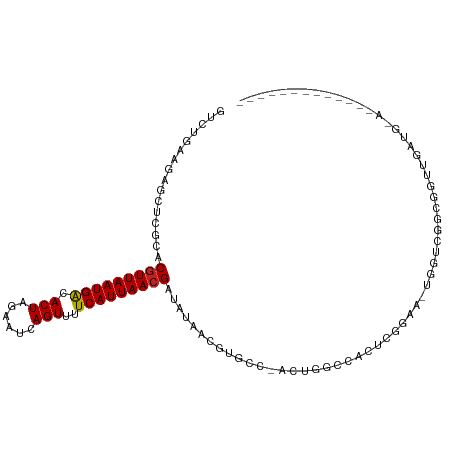
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:12:07 2011