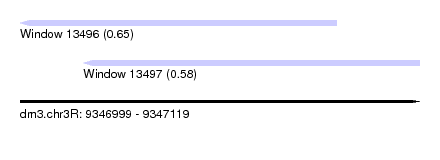
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,346,999 – 9,347,119 |
| Length | 120 |
| Max. P | 0.654609 |

| Location | 9,346,999 – 9,347,094 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 86.82 |
| Shannon entropy | 0.25093 |
| G+C content | 0.45173 |
| Mean single sequence MFE | -20.75 |
| Consensus MFE | -17.80 |
| Energy contribution | -17.90 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.654609 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
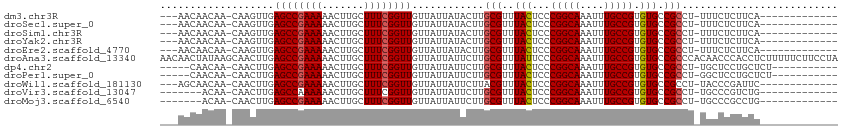
>dm3.chr3R 9346999 95 - 27905053 ---AACAACAA-CAAGUUGAGCCGAAAAACUUGCUUUCGGUUGUUAUUAUACUUGCGUUUACUCCCGGCAAAUUUGCCGUGUGCCGCCU-UUUCUCUUCA------------- ---..((((..-...))))((((((((.......))))))))............(((..(((...(((((....))))).))).)))..-..........------------- ( -21.60, z-score = -1.58, R) >droSec1.super_0 8460603 95 + 21120651 ---AACAACAA-CAAGUUGAGCCGAAAAACUUGCUUUCGGUUGUUAUUAUACUUGCGUUUACUCCCGGCAAAUUUGCCGUGUGCCGCCU-UUUCUCUUCA------------- ---..((((..-...))))((((((((.......))))))))............(((..(((...(((((....))))).))).)))..-..........------------- ( -21.60, z-score = -1.58, R) >droSim1.chr3R 15440750 95 + 27517382 ---AACAACAA-CAAGUUGAGCCGAAAAACUUGCUUUCGGUUGUUAUUAUACUUGCGUUUACUCCCGGCAAAUUUGCCGUGUGCCGCCU-UUUCUCUUCA------------- ---..((((..-...))))((((((((.......))))))))............(((..(((...(((((....))))).))).)))..-..........------------- ( -21.60, z-score = -1.58, R) >droYak2.chr3R 13653827 95 - 28832112 ---AACAACAA-CAAGUUGAGCCGAAAAACUUGCUUUCGGUUGUUAUUAUACUUGCGUUUACUCCCGGCAAAUUUGCCGUGUGCCGCCU-UUUCUCUUCA------------- ---..((((..-...))))((((((((.......))))))))............(((..(((...(((((....))))).))).)))..-..........------------- ( -21.60, z-score = -1.58, R) >droEre2.scaffold_4770 5450410 95 + 17746568 ---AACAACAA-CAAGUUGAGCCGAAAAACUUGCUUUCGGUUGUUAUUAUACUUGCGUUUACUCCCGGCAAAUUUGCCGUGUGCCGCCU-UUUCUCUUCA------------- ---..((((..-...))))((((((((.......))))))))............(((..(((...(((((....))))).))).)))..-..........------------- ( -21.60, z-score = -1.58, R) >droAna3.scaffold_13340 1900638 113 - 23697760 AACAACUAUAAGCAACUUGAGCCGAAAAACUUGCUUUCGGUUGUUAUUAUUCUUGCGUUUAUUCCCGGCAAAUUUGCCGUGUGCCGCCCACAACCCACCUCUUUUUCUUCCUA (((((((..((((((.((.........)).))))))..))))))).........(((..(((...(((((....))))).))).))).......................... ( -21.50, z-score = -1.68, R) >dp4.chr2 16789085 95 - 30794189 -----CAACAA-CAACUUGAGCCGAAAAACUUGCUUUCGGUUGUUAUUAUUCUUGCGUUUACUCCCGGCAAAUUUGCCGUGUGCCGCCU-UGCUCCUGCUCU----------- -----....((-(((((.((((..........))))..))))))).........(((..(((...(((((....))))).))).)))..-.((....))...----------- ( -20.50, z-score = -1.02, R) >droPer1.super_0 8218037 95 + 11822988 -----CAACAA-CAACUUGAGCCGAAAAACUUGCUUUCGGUUGUUAUUAUUCUUGCGUUUACUCCCGGCAAAUUUGCCGUGUGCCGCCU-GGCUCCUGCUCU----------- -----....((-(((((.((((..........))))..))))))).........(((..(((...(((((....))))).))).)))..-(((....)))..----------- ( -21.20, z-score = -0.62, R) >droWil1.scaffold_181130 6416165 95 - 16660200 ---AGCAACAA-CAACUUGAGCCGAAAAACUUGCUUUCGGUUGUUAUUAUUCUUACGUUUACUCCCGGCAAAUUUGCCGUGUGCCGCCU-UACCCGAUUC------------- ---.(((..((-(((((.((((..........))))..)))))))....................(((((....)))))..))).....-..........------------- ( -18.00, z-score = -0.52, R) >droVir3.scaffold_13047 9139089 91 - 19223366 -------ACAA-CAACUUGAGCCAAAAAACUUGCUUUCGGUUGUUAUUAUUCUUGCGUUUACUCCCGGCAAAUUUGCCGUGUGCCGCCU-UGCCCGUCUG------------- -------..((-(((((.((((..........))))..))))))).........(((..(((...(((((....))))).))).)))..-..........------------- ( -19.50, z-score = -1.08, R) >droMoj3.scaffold_6540 2642969 91 + 34148556 -------ACAA-CAACUUGAGCCGAAAAACUUGCUUUCGGUUGUUAUUAUUCUUGCGUUUACUCCCGGCAAAUUUGCCGUGUGCCGCCU-UGCCCGCCUG------------- -------..((-(((((.((((..........))))..))))))).........(((..(((...(((((....))))).))).)))..-..........------------- ( -19.50, z-score = -0.71, R) >consensus ___AACAACAA_CAACUUGAGCCGAAAAACUUGCUUUCGGUUGUUAUUAUUCUUGCGUUUACUCCCGGCAAAUUUGCCGUGUGCCGCCU_UUCCCCUUCA_____________ ...................((((((((.......))))))))............(((..(((...(((((....))))).))).))).......................... (-17.80 = -17.90 + 0.10)

| Location | 9,347,018 – 9,347,119 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 88.75 |
| Shannon entropy | 0.19140 |
| G+C content | 0.42452 |
| Mean single sequence MFE | -21.50 |
| Consensus MFE | -16.38 |
| Energy contribution | -16.98 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.582604 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
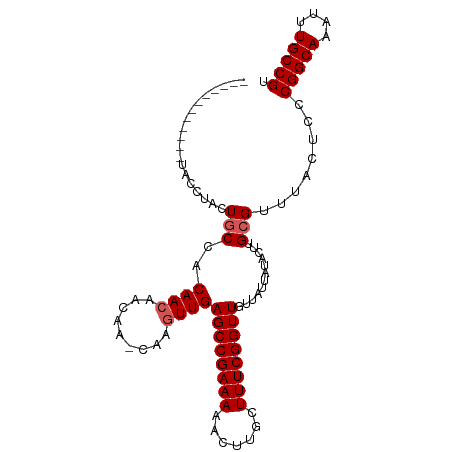
>dm3.chr3R 9347018 101 - 27905053 CAGCAGCGGCAGAACCUACUGCCACAACAACAA-CAAGUUGAGCCGAAAAACUUGCUUUCGGUUGUUAUUAUACUUGCGUUUACUCCCGGCAAAUUUGCCGU ..((((.(((((......))))).)..((((..-...))))((((((((.......))))))))...........))).........(((((....))))). ( -29.50, z-score = -2.36, R) >droSim1.chr3R 15440769 89 + 27517382 ------------UACCUACUGCCACAACAACAA-CAAGUUGAGCCGAAAAACUUGCUUUCGGUUGUUAUUAUACUUGCGUUUACUCCCGGCAAAUUUGCCGU ------------........((.....((((..-...))))((((((((.......))))))))............)).........(((((....))))). ( -19.00, z-score = -1.34, R) >droYak2.chr3R 13653846 89 - 28832112 ------------UACCUACUGCCACAACAACAA-CAAGUUGAGCCGAAAAACUUGCUUUCGGUUGUUAUUAUACUUGCGUUUACUCCCGGCAAAUUUGCCGU ------------........((.....((((..-...))))((((((((.......))))))))............)).........(((((....))))). ( -19.00, z-score = -1.34, R) >droEre2.scaffold_4770 5450429 89 + 17746568 ------------UACCUACUGCCACAACAACAA-CAAGUUGAGCCGAAAAACUUGCUUUCGGUUGUUAUUAUACUUGCGUUUACUCCCGGCAAAUUUGCCGU ------------........((.....((((..-...))))((((((((.......))))))))............)).........(((((....))))). ( -19.00, z-score = -1.34, R) >droAna3.scaffold_13340 1900671 93 - 23697760 ---------AAACUCAAACUACAACAACUAUAAGCAACUUGAGCCGAAAAACUUGCUUUCGGUUGUUAUUAUUCUUGCGUUUAUUCCCGGCAAAUUUGCCGU ---------((((.(((.....(((((((..((((((.((.........)).))))))..))))))).......))).)))).....(((((....))))). ( -21.00, z-score = -2.66, R) >consensus ____________UACCUACUGCCACAACAACAA_CAAGUUGAGCCGAAAAACUUGCUUUCGGUUGUUAUUAUACUUGCGUUUACUCCCGGCAAAUUUGCCGU ...................(((.....((((......))))((((((((.......))))))))............)))........(((((....))))). (-16.38 = -16.98 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:12:04 2011