
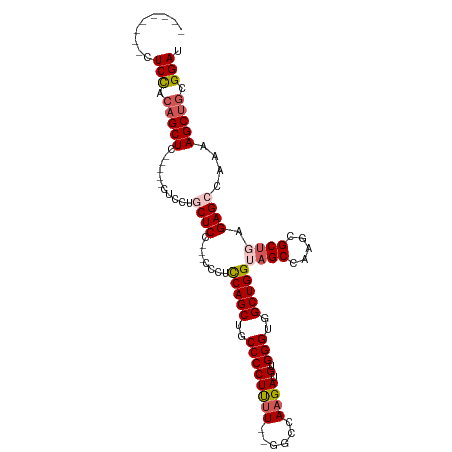
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,332,329 – 9,332,432 |
| Length | 103 |
| Max. P | 0.556918 |

| Location | 9,332,329 – 9,332,432 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 85.74 |
| Shannon entropy | 0.22967 |
| G+C content | 0.62098 |
| Mean single sequence MFE | -39.74 |
| Consensus MFE | -29.44 |
| Energy contribution | -29.96 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.556918 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9332329 103 + 27905053 CUCCACAGCUCUACAGCUC-----CUCCUGCUCC---CCCUCCAGCUGCCCCUUUUUUGGCCAAGAUGUGGGUGGCUGGUAGCCAAGCGCUGAGAGCCAAAAGCUGCGGAU .(((.(((((.....((((-----.((.((((..---.(..(((((..(((...((((....))))...)))..)))))..)...))))..))))))....))))).))). ( -40.70, z-score = -1.70, R) >droSim1.chr3R 15418533 93 - 27517382 --------CUCCACAGCUC-----CUCCUGCUCC---CCCUCCAGCUGCCCCUUUU--GGCCAAGAUGUGGGUGGCUGGUAGCCAAGCGCUGAGAGCCAAAAGCUGCGGAU --------.(((.(((((.-----.....((((.---....(((((..((((((((--....)))).).)))..)))))((((.....)))).))))....))))).))). ( -37.20, z-score = -1.42, R) >droSec1.super_0 8447665 93 - 21120651 --------CUCCGCAGCUC-----CUCCUGCUCC---CCCUCCAGCUGCCCCUUCU--GGCCAAGAUGUGGGUGGCUGGUAGCCAAGCGCUGAGAGCUAAAAGCUGCGGAU --------.(((((((((.-----.....((((.---....(((((..((((.(((--.....))).).)))..)))))((((.....)))).))))....))))))))). ( -45.80, z-score = -3.84, R) >droYak2.chr3R 13640996 98 + 28832112 --------CUCCACAGCUCUGCAGCUCCUCCUCC---ACCUUCAGCUGCCCCUUUU--GGCCAAGAUGUGGGUGGCUGGAAGCCAAGCGCCGAGAGCCGAAAGCUGCGGAU --------.(((.(((((...(.((((.((....---...((((((..((((((((--....)))).).)))..)))))).((.....)).)))))).)..))))).))). ( -36.50, z-score = -0.33, R) >droEre2.scaffold_4770 5438220 98 - 17746568 --------CUCCACAGCUCUGCAGCUCCUCCUCCACCUCCUCCAGCUGCCCCUCUU--GGCCAAGAUGUGGGUGGCUGGAAGCCAAGCGCUGAGAGCCAAAAGC---GGAU --------.(((.(.(((((.((((...............((((((..((((((((--....)))).).)))..)))))).((...))))))))))).....).---))). ( -38.50, z-score = -1.23, R) >consensus ________CUCCACAGCUC_____CUCCUGCUCC___CCCUCCAGCUGCCCCUUUU__GGCCAAGAUGUGGGUGGCUGGUAGCCAAGCGCUGAGAGCCAAAAGCUGCGGAU .........(((.(((((......((((.((..........(((((..((((((((......)))).).)))..)))))..((...)))).).))).....))))).))). (-29.44 = -29.96 + 0.52)


| Location | 9,332,329 – 9,332,432 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 85.74 |
| Shannon entropy | 0.22967 |
| G+C content | 0.62098 |
| Mean single sequence MFE | -46.44 |
| Consensus MFE | -31.12 |
| Energy contribution | -32.24 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.524862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9332329 103 - 27905053 AUCCGCAGCUUUUGGCUCUCAGCGCUUGGCUACCAGCCACCCACAUCUUGGCCAAAAAAGGGGCAGCUGGAGGG---GGAGCAGGAG-----GAGCUGUAGAGCUGUGGAG .(((((((((((((((((((...(((((((.....))).(((...(((..(((........)))....))).))---)))))..)).-----)))))).))))))))))). ( -48.10, z-score = -2.04, R) >droSim1.chr3R 15418533 93 + 27517382 AUCCGCAGCUUUUGGCUCUCAGCGCUUGGCUACCAGCCACCCACAUCUUGGCC--AAAAGGGGCAGCUGGAGGG---GGAGCAGGAG-----GAGCUGUGGAG-------- .((((((((((((.(((((((((.....))).(((((..(((....(((....--..))))))..)))))...)---)))))...))-----)))))))))).-------- ( -44.10, z-score = -1.85, R) >droSec1.super_0 8447665 93 + 21120651 AUCCGCAGCUUUUAGCUCUCAGCGCUUGGCUACCAGCCACCCACAUCUUGGCC--AGAAGGGGCAGCUGGAGGG---GGAGCAGGAG-----GAGCUGCGGAG-------- .((((((((((((.(((((((((.....))).(((((..(((.(.(((.....--))).))))..)))))...)---)))))...))-----)))))))))).-------- ( -48.00, z-score = -3.13, R) >droYak2.chr3R 13640996 98 - 28832112 AUCCGCAGCUUUCGGCUCUCGGCGCUUGGCUUCCAGCCACCCACAUCUUGGCC--AAAAGGGGCAGCUGAAGGU---GGAGGAGGAGCUGCAGAGCUGUGGAG-------- .((((((((((((((((((...(.(((.((((.((((..(((....(((....--..))))))..)))).))))---.)))).))))))).))))))))))).-------- ( -47.60, z-score = -2.00, R) >droEre2.scaffold_4770 5438220 98 + 17746568 AUCC---GCUUUUGGCUCUCAGCGCUUGGCUUCCAGCCACCCACAUCUUGGCC--AAGAGGGGCAGCUGGAGGAGGUGGAGGAGGAGCUGCAGAGCUGUGGAG-------- .(((---((....((((((((((.(((.(((((((((..(((...((((....--)))).)))..))))))...))).))).....)))).))))))))))).-------- ( -44.40, z-score = -0.76, R) >consensus AUCCGCAGCUUUUGGCUCUCAGCGCUUGGCUACCAGCCACCCACAUCUUGGCC__AAAAGGGGCAGCUGGAGGG___GGAGCAGGAG_____GAGCUGUGGAG________ .((((((((((....(((.(.((.((...((.(((((..(((....(((........))))))..))))).))....)).)).)))).....))))))))))......... (-31.12 = -32.24 + 1.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:12:01 2011