| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,331,471 – 9,331,590 |
| Length | 119 |
| Max. P | 0.518279 |

| Location | 9,331,471 – 9,331,590 |
|---|---|
| Length | 119 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.58 |
| Shannon entropy | 0.43390 |
| G+C content | 0.30942 |
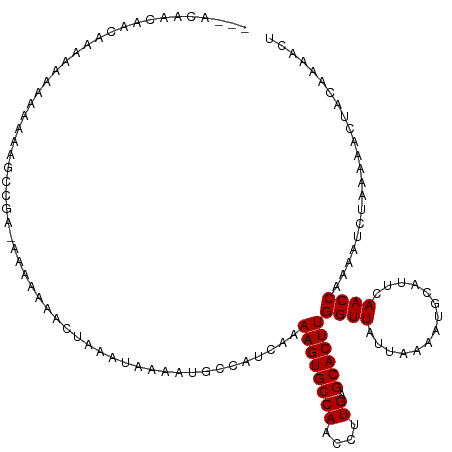
| Mean single sequence MFE | -12.33 |
| Consensus MFE | -9.04 |
| Energy contribution | -9.04 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.79 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.518279 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 9331471 119 + 27905053 ACUGCAACAAAAAAAAAAUA-GAGCCGAAAAAAAAACUAAAUAAAAUGCCAUCAAAAGUGCCAACCUUGAGCACUUGGUUAUUAAAAUGCAUUCAACCAAAAUCUAAAAACUACAAAACU ..................((-((................................((((((((....)).))))))((((..............))))....)))).............. ( -11.44, z-score = -0.57, R) >droSim1.chr3R 15417694 117 - 27517382 ---ACUACAAAAAAAAAAAAUGAGUCGAAAAAAAAACUAAAUAAAAUGCCAUCAAAAGUGCCAACCUUGAGCACUUGGUUAUUAAAAUGCAUUCAACCAAAAUCUAAAAACUACAAAACU ---.................(((((..............................((((((((....)).))))))((((..............))))...........))).))..... ( -9.94, z-score = -0.67, R) >droSec1.super_0 8446806 120 - 21120651 ACUGCAAAAAAAAAAAAAAAUGAGUCGAAAAAAAAACUAAAUAAAAUGCCAUCAAAAGUGCCAACCUUGAGCACUUGGUUAUUAAAAUGCAUUCAACCAAAAUCUAAAAACUACAAAACU ..((((................(((..........))).........((((.....(((((((....)).)))))))))........))))............................. ( -11.20, z-score = -0.75, R) >droYak2.chr3R 13640090 111 + 28832112 ---------ACUACAAAAAAAAGCUAGAAAAAAAAACUAAAUAAAAUGCCAUCAAAAGUGCCAACCUUGAGCACUUGGUUAUUAAAAUGCAUUCAACCAAAAUCUAAAAACUACAAAACU ---------...............((((...........................((((((((....)).))))))((((..............))))....)))).............. ( -11.24, z-score = -1.22, R) >droEre2.scaffold_4770 5437347 115 - 17746568 ---ACUACAACAAAAAUAACAAAGUUG--AAAAAAACUAAAUAAAAUGCCAUCAAAAGUGCCAACCUUGAGCACUUGGUUAUUAAAAUGCAUUCAACCAAAAUCUAAAAACUACAAAACU ---...........((((((..((((.--.....)))).................((((((((....)).)))))).))))))..................................... ( -13.20, z-score = -1.72, R) >droAna3.scaffold_13340 1888098 112 + 23697760 ---AUGCCGGCAACAAAAACAAAAA-----AAAAAACUGAAUAAAAUGCCAUCAAAAGUGCCAACCUUGAGCACUUGGUUAUUAAAAUGCAUUCAACCAAAAUCUAAAAACUACAAAACU ---..(((((((.............-----................)))).....((((((((....)).)))))))))......................................... ( -13.65, z-score = -0.93, R) >dp4.chr2 16777328 113 + 30794189 ---ACAGCAGCAGCAACAAAAAAGCU----AAAAAACUAAAUAAAAUGCCAUCAAAAGUGCCAACCUUGAGCACUUGGUUAUUAAAAUGCAUUCAACCAAAAUCUAAAAACUACAAAACU ---...(((..(((.........)))----.................((((.....(((((((....)).)))))))))........))).............................. ( -13.30, z-score = -0.17, R) >droPer1.super_0 8206264 113 - 11822988 ---ACAACAGCAGCAACAAAAAAGCU----AAAAAACUAAAUAAAAUGCCAUCAAAAGUGCCAACCUUGAGCACUUGGUUAUUAAAAUGCAUUCAACCAAAAUCUAAAAACUACAAAACU ---......(((((.........)))----.................((((.....(((((((....)).))))))))).........)).............................. ( -12.30, z-score = -0.41, R) >droWil1.scaffold_181130 6402954 120 + 16660200 AGAACAAAAAAUAUAUAAAAUAAAACGGCAAAAAAUGGAAGAGGAAUGCCAUCAAAAGUGCCAACCUUGAGCACUUGGUUAUUAAAAUGCAUUCAACCAAAAUCUAAAAACUACAAAACU ...................................(((.....((((((......((((((((....)).)))))).(((.....)))))))))..)))..................... ( -14.80, z-score = -0.46, R) >droVir3.scaffold_13047 9128645 93 + 19223366 ---------------------------ACACGCACAGAGACGAAAAUUUCAUCAAAAGUGCCAACCUUGAGCACUUGGUUAUUAAAAUGCAUUCAACCAAAAUCUAAAAACUACCAAACU ---------------------------....(((..((((......)))).....((((((((....)).))))))...........))).............................. ( -11.60, z-score = -1.05, R) >droGri2.scaffold_14906 10054707 103 - 14172833 -----------------ACAGGCACAGACGCACCCACUCACGAAAAUUCCAUCAAAAGUGCCAACCUUGAGCACUUGGUUAUUAAAAUGCAUUCAACCAACAUCAAAAAACUACCAAACU -----------------...(((((.((.(......)))..((........))....)))))....((((....((((((..............))))))..)))).............. ( -12.94, z-score = -0.72, R) >consensus ___ACAACAACAAAAAAAAAAAAGCCGA_AAAAAAACUAAAUAAAAUGCCAUCAAAAGUGCCAACCUUGAGCACUUGGUUAUUAAAAUGCAUUCAACCAAAAUCUAAAAACUACAAAACU .......................................................((((((((....)).))))))((((..............))))...................... ( -9.04 = -9.04 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:12:00 2011