
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,329,435 – 9,329,553 |
| Length | 118 |
| Max. P | 0.967676 |

| Location | 9,329,435 – 9,329,553 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 77.13 |
| Shannon entropy | 0.41241 |
| G+C content | 0.53312 |
| Mean single sequence MFE | -41.49 |
| Consensus MFE | -25.81 |
| Energy contribution | -26.13 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.967676 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
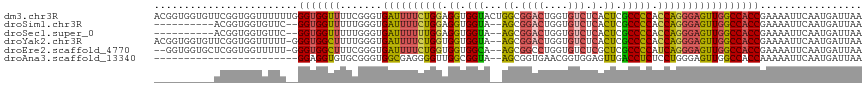
>dm3.chr3R 9329435 118 + 27905053 ACGGUGGUGUUCGGUGGUUUUUUGGGUGGUUUUCGGGUGAUUUUCUGGAGGUGGUACUGGCGGACUGGUGUCUCACUCGCCCCACCAGGGAGUUGGCCACCGAAAAUUCAAUGAUUAA .((.(((..((((((((((.((((((.....)))))).((((..((((.((.(((...(..((((....))))..)..))))).))))..))))))))))))))...))).))..... ( -40.60, z-score = -0.69, R) >droSim1.chr3R 15415540 104 - 27517382 ----------ACGGUGGUGUUC--GGUGGUUUUUGGGUGAUUUUCUGGAGGUGGUA--AGCGGACUGGUGUCUCACUCGCCCCACCAGGGAGUUGGCCACCGAAAAUUCAAUGAUUAA ----------.((.(((..(((--(((((((.......((((..((((.((.((..--((.((((....))).).))..)))).))))..))))))))))))))...))).))..... ( -41.51, z-score = -2.55, R) >droSec1.super_0 8444325 104 - 21120651 ----------ACGGUGGUGUUC--GGUGGUUUUUGGGUGAUUUUUUGGAGGUGGUA--AGCGGACUGGUGUCUCACUCGCCCCACCAGGGAGUUGGCCACCGAAAAUUCAAUGAUUAA ----------.((.(((..(((--(((((((.......((((((((((.((.((..--((.((((....))).).))..)))).))))))))))))))))))))...))).))..... ( -38.81, z-score = -2.01, R) >droYak2.chr3R 13638049 115 + 28832112 ACGGUGGUGUUCGGUGGUUUUU-GGGUGGCUUUUGGGUGAUUUUCUGGUGGUGGUA--AGCGGACUGGUGUCUCACUCGCCCCACCAGGGAGUUGGCCACCGAAAAUUCAAUGAUUAA ..........(((.((..((((-.(((((((.......((((..(((((((.((..--((.((((....))).).))..)))))))))..))))))))))).))))..)).))).... ( -48.11, z-score = -3.36, R) >droEre2.scaffold_4770 5435372 113 - 17746568 --GGUGGUGCUCGGUGGUUUUU-GGGUGGCUUUCGGGUGAUUUUCUGGUGGUGGCA--AGCGGCCUGGUGUCUCGCUCGCCCCAUCAGGGAGUUGGCCACCGAAAAUUCAAUGAUUAA --((((((((((((.((((...-....)))).))))))((((..(((((((.(((.--(((((.(....).).)))).))))))))))..)))).))))))................. ( -48.00, z-score = -2.83, R) >droAna3.scaffold_13340 1886315 92 + 23697760 ------------------------GGAGGUGUGCGGGUGGCGAGGGGUUGGCGGUA--AGCGGUGAACGGUGGAGUUGACCUCUCCUGGGAGUUGGCCACCAAAAAUUCAAUGAUUAA ------------------------.(((.......(((((((((((((..((....--.((.(....).))...))..)))))))(........))))))).....)))......... ( -31.90, z-score = -1.55, R) >consensus __________UCGGUGGUUUUU__GGUGGUUUUCGGGUGAUUUUCUGGAGGUGGUA__AGCGGACUGGUGUCUCACUCGCCCCACCAGGGAGUUGGCCACCGAAAAUUCAAUGAUUAA ........................(((((((.......((((((((((.((.(((...((.((((....))).).)).))))).)))))))))))))))))................. (-25.81 = -26.13 + 0.31)

| Location | 9,329,435 – 9,329,553 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 77.13 |
| Shannon entropy | 0.41241 |
| G+C content | 0.53312 |
| Mean single sequence MFE | -30.10 |
| Consensus MFE | -15.45 |
| Energy contribution | -16.84 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.966701 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
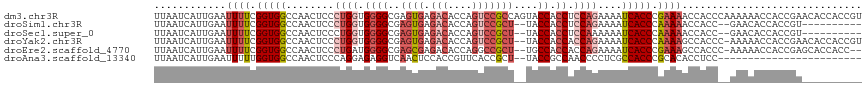
>dm3.chr3R 9329435 118 - 27905053 UUAAUCAUUGAAUUUUCGGUGGCCAACUCCCUGGUGGGGCGAGUGAGACACCAGUCCGCCAGUACCACCUCCAGAAAAUCACCCGAAAACCACCCAAAAAACCACCGAACACCACCGU ..............((((((((..........(((((..(..(((.(((....))))))..)..))))).........((....))...............))))))))......... ( -29.70, z-score = -1.47, R) >droSim1.chr3R 15415540 104 + 27517382 UUAAUCAUUGAAUUUUCGGUGGCCAACUCCCUGGUGGGGCGAGUGAGACACCAGUCCGCU--UACCACCUCCAGAAAAUCACCCAAAAACCACC--GAACACCACCGU---------- ..............((((((((........((((.((((..((((.(((....)))))))--..)).)).))))...............)))))--))).........---------- ( -31.20, z-score = -2.63, R) >droSec1.super_0 8444325 104 + 21120651 UUAAUCAUUGAAUUUUCGGUGGCCAACUCCCUGGUGGGGCGAGUGAGACACCAGUCCGCU--UACCACCUCCAAAAAAUCACCCAAAAACCACC--GAACACCACCGU---------- ..............(((((((((((......))).((((..((((.(((....)))))))--..)).))....................)))))--))).........---------- ( -28.60, z-score = -2.07, R) >droYak2.chr3R 13638049 115 - 28832112 UUAAUCAUUGAAUUUUCGGUGGCCAACUCCCUGGUGGGGCGAGUGAGACACCAGUCCGCU--UACCACCACCAGAAAAUCACCCAAAAGCCACCC-AAAAACCACCGAACACCACCGU ..............((((((((........(((((((((..((((.(((....)))))))--..)).)))))))..............(.....)-.....))))))))......... ( -36.30, z-score = -4.05, R) >droEre2.scaffold_4770 5435372 113 + 17746568 UUAAUCAUUGAAUUUUCGGUGGCCAACUCCCUGAUGGGGCGAGCGAGACACCAGGCCGCU--UGCCACCACCAGAAAAUCACCCGAAAGCCACCC-AAAAACCACCGAGCACCACC-- ..............((((((((........(((.(((((((((((.(........)))))--)))).))).))).........(....)......-.....)))))))).......-- ( -35.00, z-score = -3.70, R) >droAna3.scaffold_13340 1886315 92 - 23697760 UUAAUCAUUGAAUUUUUGGUGGCCAACUCCCAGGAGAGGUCAACUCCACCGUUCACCGCU--UACCGCCAACCCCUCGCCACCCGCACACCUCC------------------------ .................((((((...(((....))).(((.(((......))).)))...--...............))))))...........------------------------ ( -19.80, z-score = -1.49, R) >consensus UUAAUCAUUGAAUUUUCGGUGGCCAACUCCCUGGUGGGGCGAGUGAGACACCAGUCCGCU__UACCACCACCAGAAAAUCACCCAAAAACCACC__AAAAACCACCGA__________ ............((((.(((((..........(((((....((((.(((....)))))))....))))).........))))).)))).............................. (-15.45 = -16.84 + 1.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:11:59 2011