
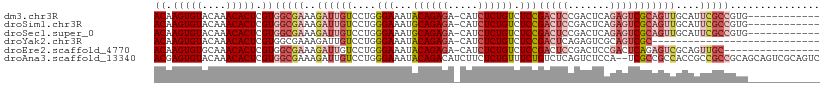
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,321,114 – 9,321,212 |
| Length | 98 |
| Max. P | 0.996627 |

| Location | 9,321,114 – 9,321,212 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 80.14 |
| Shannon entropy | 0.35885 |
| G+C content | 0.52046 |
| Mean single sequence MFE | -33.40 |
| Consensus MFE | -21.94 |
| Energy contribution | -24.67 |
| Covariance contribution | 2.72 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.976400 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
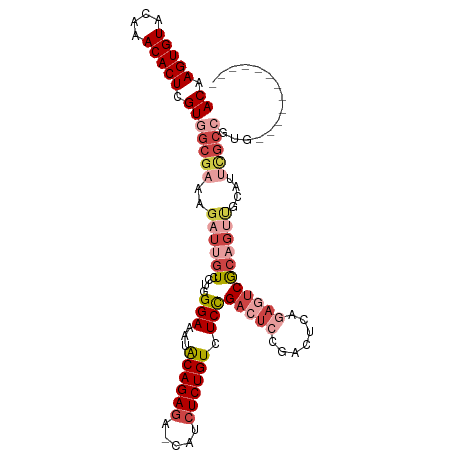
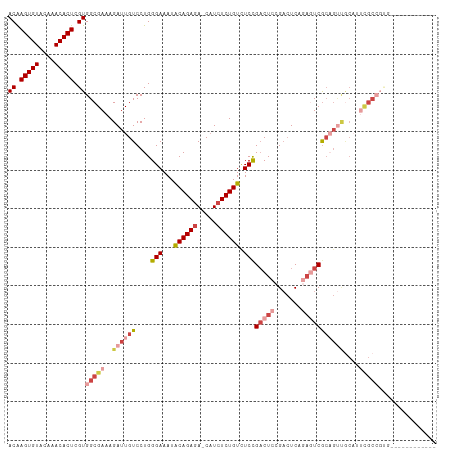
>dm3.chr3R 9321114 98 + 27905053 ACAAGUGUACAAACACUCGUGGCGAAAGAUUGUCCUGGGAAAUACAGAGA-CAUCUCUGUCUCCGACUCCGACUCAGAGUCGCAGUUGCAUUCGCCGUG------------ ((.(((((....))))).))((((((.((((((....(((...((((((.-...)))))).)))(((((.......)))))))))))...))))))...------------ ( -36.00, z-score = -3.20, R) >droSim1.chr3R 15407237 98 - 27517382 ACAAGUGUACAAACACUCGUGGCGAAAGAUUGUCCUGGGAAAUGCAGAGA-CAUCUCUGUCUCCGACUCCGACUCAGAGUCGCAGUUGCAUUCGCCGUG------------ ((.(((((....))))).))((((((.((((((....(((...((((((.-...)))))).)))(((((.......)))))))))))...))))))...------------ ( -36.30, z-score = -2.88, R) >droSec1.super_0 8436205 98 - 21120651 ACAAGUGUACAAACACUCGUGGCGAAAGAUUGUCCUGGGAAAUGCAGAGA-CAUCUCUGUCUCCGACUCCGACUCAGAGUCGCAGUUGCAUUCGCCGUG------------ ((.(((((....))))).))((((((.((((((....(((...((((((.-...)))))).)))(((((.......)))))))))))...))))))...------------ ( -36.30, z-score = -2.88, R) >droYak2.chr3R 13629712 82 + 28832112 ACAAGUGUACAAACACUCGUGGCGAAAGAUUGUCCUGGGAAAUACAGAGA-CAUCUCUGUCUCCGACUCAGAGUCGCAGUCGC---------------------------- ((.(((((....))))).)).......((((((....(((...((((((.-...)))))).)))(((.....)))))))))..---------------------------- ( -24.40, z-score = -1.27, R) >droEre2.scaffold_4770 5427261 94 - 17746568 ACAAGUGUGCAAACACUCGUGGCGAAAGAUUGUCCUGGGAAAUACAGAGA-CAUCUCUGUCUCCGACUCCGACUCCGACUCAGAGUCGCAGUUGC---------------- ((.(((((....))))).))((.((.(((.(((((((.......))).))-))..))).)).))((((.((((((.......)))))).))))..---------------- ( -31.20, z-score = -1.69, R) >droAna3.scaffold_13340 1878124 109 + 23697760 ACGAGUGUACAAACACUCGUGGCGAAAGAUUGUCCUGGGAAAUACAGACAUCUUCUCUGUUUCUGUCUCAGUCUCCA--UCGCCGCCACCGCCGCCGCAGCAGUCGCAGUC ((((((((....))))))))(((((.((((((.....(((((((.(((.....))).)))))))....))))))...--)))))((.((.((.......)).)).)).... ( -36.20, z-score = -2.55, R) >consensus ACAAGUGUACAAACACUCGUGGCGAAAGAUUGUCCUGGGAAAUACAGAGA_CAUCUCUGUCUCCGACUCCGACUCAGAGUCGCAGUUGCAUUCGCCGUG____________ ((.(((((....))))).))(((((..((((((....(((...((((((.....)))))).)))(((((.......)))))))))))....)))))............... (-21.94 = -24.67 + 2.72)
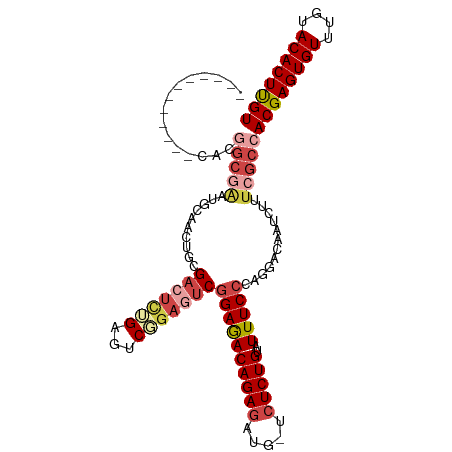
| Location | 9,321,114 – 9,321,212 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 80.14 |
| Shannon entropy | 0.35885 |
| G+C content | 0.52046 |
| Mean single sequence MFE | -38.47 |
| Consensus MFE | -25.45 |
| Energy contribution | -27.45 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.96 |
| SVM RNA-class probability | 0.996627 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9321114 98 - 27905053 ------------CACGGCGAAUGCAACUGCGACUCUGAGUCGGAGUCGGAGACAGAGAUG-UCUCUGUAUUUCCCAGGACAAUCUUUCGCCACGAGUGUUUGUACACUUGU ------------...((((((.....((.((((((((...)))))))).))((((((...-.)))))).................))))))((((((((....)))))))) ( -40.60, z-score = -3.78, R) >droSim1.chr3R 15407237 98 + 27517382 ------------CACGGCGAAUGCAACUGCGACUCUGAGUCGGAGUCGGAGACAGAGAUG-UCUCUGCAUUUCCCAGGACAAUCUUUCGCCACGAGUGUUUGUACACUUGU ------------...((((((((((....((((((((...))))))))((((((....))-))))))))..((....))......))))))((((((((....)))))))) ( -41.00, z-score = -3.72, R) >droSec1.super_0 8436205 98 + 21120651 ------------CACGGCGAAUGCAACUGCGACUCUGAGUCGGAGUCGGAGACAGAGAUG-UCUCUGCAUUUCCCAGGACAAUCUUUCGCCACGAGUGUUUGUACACUUGU ------------...((((((((((....((((((((...))))))))((((((....))-))))))))..((....))......))))))((((((((....)))))))) ( -41.00, z-score = -3.72, R) >droYak2.chr3R 13629712 82 - 28832112 ----------------------------GCGACUGCGACUCUGAGUCGGAGACAGAGAUG-UCUCUGUAUUUCCCAGGACAAUCUUUCGCCACGAGUGUUUGUACACUUGU ----------------------------((((((.((((.....)))).))...((((((-((.(((.......)))))).))))))))).((((((((....)))))))) ( -29.40, z-score = -2.48, R) >droEre2.scaffold_4770 5427261 94 + 17746568 ----------------GCAACUGCGACUCUGAGUCGGAGUCGGAGUCGGAGACAGAGAUG-UCUCUGUAUUUCCCAGGACAAUCUUUCGCCACGAGUGUUUGCACACUUGU ----------------...(((.((((((((...)))))))).))).((.((.(((..((-((.(((.......))))))).))).)).))((((((((....)))))))) ( -35.90, z-score = -2.22, R) >droAna3.scaffold_13340 1878124 109 - 23697760 GACUGCGACUGCUGCGGCGGCGGUGGCGGCGA--UGGAGACUGAGACAGAAACAGAGAAGAUGUCUGUAUUUCCCAGGACAAUCUUUCGCCACGAGUGUUUGUACACUCGU ....((.(((((((...))))))).))(((((--.(((..(((.((..((.(((((.......))))).)))).))).....))).)))))((((((((....)))))))) ( -42.90, z-score = -2.63, R) >consensus ____________CACGGCGAAUGCAACUGCGACUCUGAGUCGGAGUCGGAGACAGAGAUG_UCUCUGUAUUUCCCAGGACAAUCUUUCGCCACGAGUGUUUGUACACUUGU ...............(((((..........(((((((...)))))))((((((((((.....)))))..)))))............)))))((((((((....)))))))) (-25.45 = -27.45 + 2.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:11:56 2011