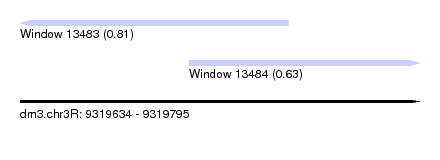
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,319,634 – 9,319,795 |
| Length | 161 |
| Max. P | 0.808702 |

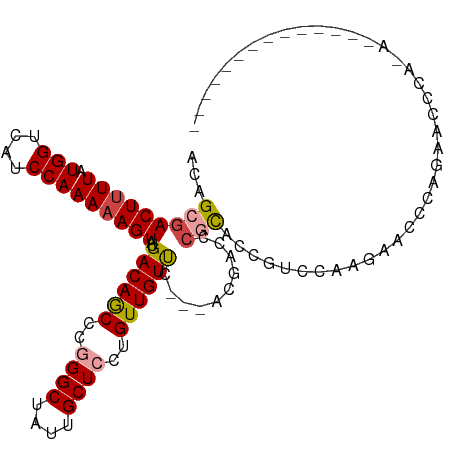
| Location | 9,319,634 – 9,319,742 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 74.88 |
| Shannon entropy | 0.43294 |
| G+C content | 0.50590 |
| Mean single sequence MFE | -20.37 |
| Consensus MFE | -16.25 |
| Energy contribution | -16.75 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.14 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.808702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9319634 108 - 27905053 ACAGCGACUUUUAUGGUCAUCCAAAAAGUAUGACAGCCCGGGCUAUUGCUCCUGUUGUUC---ACGACCCGCACCGUCCAAGAACCCAGAACCCAAAACCCACAGCCCACA ...(((((((((.(((....)))))))))..((((((..((((....))))..)))))).---......)))....................................... ( -20.90, z-score = -0.57, R) >droSim1.chr3R 15405745 94 + 27517382 ACAGCGACUUUUAUGGUCAUCCAAAAAGUAUGACAGCCCGGGCUAUUGCUCCUGUUGUUC---ACGACCCGCACCGUCCAAGAGCCCAGAACCCACA-------------- ...(((((((((.(((....)))))))))..((((((..((((....))))..)))))).---......))).........................-------------- ( -20.90, z-score = -0.60, R) >droSec1.super_0 8434723 94 + 21120651 ACAGAGACUUUUAUGGUCAUCCAAAUAGUAUGACAGCCCGGGCUAUUGCUCCUGUUGUUC---ACGACCCGCACCGUCCAAGACCCCAGAACCCACA-------------- .....(..((((..((((.........((..((((((..((((....))))..)))))).---))(((.......)))...))))..))))..)...-------------- ( -18.00, z-score = -0.08, R) >droYak2.chr3R 13628089 102 - 28832112 ACAGCGACUUUUAUGGUCAUCCAAAAAGUAUGACAGCCCGGGCUAUUGCUCCUGUUGUUC---ACGACCCGCACCGUCC-AGAACCCACAGCCCAGUGCCCACCGC----- ...(((((((((.(((....)))))))))..........((((.((((...((((.((((---..(((.......))).-.))))..))))..))))))))..)))----- ( -27.30, z-score = -1.77, R) >droAna3.scaffold_13340 1877104 84 - 23697760 ACAGCGACUUUUAUGGUCAUCCAAAAAGUAUGACAACUCAGGCUACUGCUCCUGUUGUUC---CCGACCAGUAACCCAC-AGUCCACA----------------------- ...(.((((....(((((...(.....)...((((((...(((....)))...)))))).---..)))))((.....))-)))))...----------------------- ( -15.40, z-score = -0.31, R) >droWil1.scaffold_181130 6390741 83 - 16660200 ACAGAGACUUUUAUGGUCAUCCAAAAAGUAUGACAACUCAGGCUAUUGCUCUUGUUGUCCUCAGCAAUUCACAACCCACAAAG---------------------------- ...(((((((((.(((....)))))))))..((((((...(((....)))...))))))))).....................---------------------------- ( -19.70, z-score = -2.58, R) >consensus ACAGCGACUUUUAUGGUCAUCCAAAAAGUAUGACAGCCCGGGCUAUUGCUCCUGUUGUUC___ACGACCCGCACCGUCCAAGAACCCAGAACCCA_A______________ ...(((((((((.(((....)))))))))..((((((..((((....))))..))))))..........)))....................................... (-16.25 = -16.75 + 0.50)

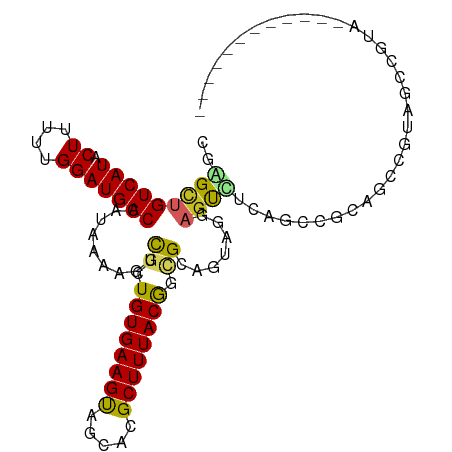
| Location | 9,319,702 – 9,319,795 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 70.21 |
| Shannon entropy | 0.61754 |
| G+C content | 0.47038 |
| Mean single sequence MFE | -25.80 |
| Consensus MFE | -12.42 |
| Energy contribution | -11.91 |
| Covariance contribution | -0.51 |
| Combinations/Pair | 1.38 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.625020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9319702 93 + 27905053 CGGGCUGUCAUACUUUUUGGAUGACCAUAAAAGUCGCUGUGAAGUAGGACGCUUUACGGCGGAGUAGGAGUUUUAGCCGUUGCCGUAGCCAUA------------ ..(((.(((.((((((.(((.((((.......))))))).)))))).)))....((((((((....((........)).)))))))))))...------------ ( -30.30, z-score = -1.57, R) >droSim1.chr3R 15405799 93 - 27517382 CGGGCUGUCAUACUUUUUGGAUGACCAUAAAAGUCGCUGUGAAGUAGGACGCUUUACGGCGGAGUAGGAGUCUUAGCCGUUGCCGUUGCCGUA------------ ..(((.(((.((((((.(((.((((.......))))))).)))))).)))((...(((((.(((.......))).))))).))....)))...------------ ( -29.60, z-score = -0.76, R) >droSec1.super_0 8434777 93 - 21120651 CGGGCUGUCAUACUAUUUGGAUGACCAUAAAAGUCUCUGUGAAGUAGGACGCUUUACGGCGGAGUAGGAGUCUUAGCCGUUGCCGUAGCCGUA------------ ..(((((((.((((...((((.(((.......)))))))...)))).)))((...(((((.(((.......))).))))).))...))))...------------ ( -28.30, z-score = -0.54, R) >droYak2.chr3R 13628151 98 + 28832112 CGGGCUGUCAUACUUUUUGGAUGACCAUAAAAGUCGCUGUGAAGUAGGACGCUUUACGGCGGAGUAGGAGUCGGAGUCUUAGCCA-AGCCGUAGCCGGA------ (.(((.(((.((((((.(((.((((.......))))))).)))))).)))....((((((((..(((((.......))))).)).-.))))))))).).------ ( -31.60, z-score = -0.75, R) >droEre2.scaffold_4770 5425722 81 - 17746568 CGAGCUGUCAUACUUUUCGGAUGACCAUAAAAGUCGCUGUGAAGUAGGACGCUUUACGGCGGAGUAGGAGUCUUAGCCGUA------------------------ .((((.(((.((((((.(((.((((.......))))))).)))))).)))))))((((((.(((.......))).))))))------------------------ ( -29.00, z-score = -2.61, R) >droAna3.scaffold_13340 1877148 98 + 23697760 UGAGUUGUCAUACUUUUUGGAUGACCAUAAAAGUCGCUGUGAAGUAGCACGCUUUACGUCGCUGUUCCU-UCAGAGCUGCUGCUGCUGCUAUUCCUGCU------ ..(((.(((((.((....)))))))......(((.(((.(((((.((((.((........)))))).))-))).))).))))))...((.......)).------ ( -24.60, z-score = 0.04, R) >droWil1.scaffold_181130 6390784 100 + 16660200 UGAGUUGUCAUACUUUUUGGAUGACCAUAAAAGUCUCUGUGAAGUAGCACGCUUUACGCUGC-ACAAACGUGUCCCACCCCGCCCUCAUCCUUUCGCCCCA---- .((((......))))...((((((..............(((((((.....)))))))(..((-((....))))..).........))))))..........---- ( -18.30, z-score = -0.50, R) >droVir3.scaffold_13047 9118618 88 + 19223366 UGAGUUGUCAUACUUUUUGGAUGACCAUAAAAGUUGCUGUGAAGUAGCACGCUUUACAUUGCCAUUGCAGCUACUUAAACAACUGAGA----------------- (.((((((.....(((.(((....))).)))((((((((((((((.....))))))))........))))))......)))))).)..----------------- ( -19.90, z-score = -0.72, R) >droMoj3.scaffold_6540 2620087 98 - 34148556 UGAGUUGUCAUACUUUUUGGAUGACCAUAAAAGUUGCUGUGAAGCAGCACGCUUUACAUUGCCAUUGCAGCUACUUAAACAACCCAAGCAACAAUAAC------- ......(((((.((....))))))).......(((((((((((((.....))))))).((((....))))................))))))......------- ( -23.10, z-score = -1.51, R) >droGri2.scaffold_14906 10044882 105 - 14172833 UGAGUUGUCAUACUUUUUGGAUGACCAUAAAAGUUGCUGUGAAGUAGCACGCUUUACAAUGCCAUUGCAGCUACUUAAACAACCAAGGCAGCAAUAACGAACAGG ...((((((((.((....))))))).......((((((((.(((((((..((..............)).)))))))...(......)))))))))....)))... ( -23.34, z-score = -0.28, R) >consensus CGAGCUGUCAUACUUUUUGGAUGACCAUAAAAGUCGCUGUGAAGUAGCACGCUUUACGGCGCAGUAGGAGUCUCAGCCGCAGCCGUAGCCGUA____________ ..(((((((((.((....))))))).........((.((((((((.....)))))))).)).......))))................................. (-12.42 = -11.91 + -0.51)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:11:54 2011