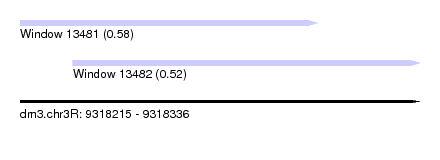
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,318,215 – 9,318,336 |
| Length | 121 |
| Max. P | 0.578739 |

| Location | 9,318,215 – 9,318,305 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 84.68 |
| Shannon entropy | 0.29261 |
| G+C content | 0.35875 |
| Mean single sequence MFE | -20.27 |
| Consensus MFE | -14.69 |
| Energy contribution | -14.87 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.578739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9318215 90 + 27905053 -------------CGUAUUAAGCCCAAAUGAUUUAAUGAGCAUGUUUUACCCAAAAUGUAGUAAUUGUCACGUAUUACGUGCUCAGGCUGAAGAUUG-CGUAUG----------- -------------((((...(((((....)......(((((((((..(((....(((......))).....)))..)))))))))))))......))-))....----------- ( -21.70, z-score = -2.05, R) >droSim1.chr3R 15403851 90 - 27517382 -------------CGUAUUAAGCCCAAAUGAUUUAAUGAGCAUGUUUUACCCAAAAUGUAGUAAUUGUCACGUAUUACGUGCUCAGGCUGAAGAUUU-CGUAUG----------- -------------.......(((((....)......(((((((((..(((....(((......))).....)))..)))))))))))))........-......----------- ( -19.20, z-score = -1.60, R) >droSec1.super_0 8433340 90 - 21120651 -------------CGUAUUAAGCCCAAAUGAUUUAAUGAGCAUGUUUUACCCAAAAUGUAGUAAUUGUCACGUAUUACGUGCUCAGGCUGAAGAUUU-CGUAUG----------- -------------.......(((((....)......(((((((((..(((....(((......))).....)))..)))))))))))))........-......----------- ( -19.20, z-score = -1.60, R) >droYak2.chr3R 13626667 89 + 28832112 -------------CGUAUUAAGCCCAAAUGAUUUAAUGAGCAUGUUUUACCCAAA-UGUAGUAAUUGUCACGUAUUACGUGCUCAGGCUGAAGAUUG-CGUAUG----------- -------------((((...(((((....)......(((((((((..(((...((-(......))).....)))..)))))))))))))......))-))....----------- ( -21.30, z-score = -1.87, R) >droEre2.scaffold_4770 5424254 90 - 17746568 -------------UGUAUUAAGCCCAAAUGAUUUAAUGAGCAUGUUUUACCCAAAAUGUAGUAAUUGUCACGUAUUACGUGCUCAGGCUGAAGAUUG-CGUAUG----------- -------------.(((...(((((....)......(((((((((..(((....(((......))).....)))..)))))))))))))......))-).....----------- ( -20.40, z-score = -1.59, R) >droAna3.scaffold_13340 1875347 92 + 23697760 -------------CAUAUUAAGCUCAAAUGAUUUAAUGAGCGUGUUUUACCCAAAAUGUAGUAAUUGUCACGUAUUACGUGCUCAGCCUGAAGAUUU-UGCUUUGA--------- -------------......((((..((((..((((.(((((((((..(((....(((......))).....)))..)))))))))...)))).))))-.))))...--------- ( -16.40, z-score = -0.42, R) >dp4.chr2 16763221 103 + 30794189 UGCCUUUUCCGUACGUAUUAAGCUCAAAUGAUUUAAUGAGCAUGUUUUACCUAAAAUGUAGUAAUUGUCACGUAUUACGUGCUCAGCCUGAAGAUUG-CGUAUG----------- .........(((((((((((((.((....)))))))(((((((((..(((....(((......))).....)))..)))))))))..........))-))))))----------- ( -24.40, z-score = -2.26, R) >droPer1.super_0 8191217 103 - 11822988 UUCCUUUGCCGUACGUAUUAAGCUCAAAUGAUUUAAUGAGCAUGUUUUACCUAAAAUGUAGUAAUUGUCACGUAUUACGUGCUCAGCCUGAAGAUUG-CGUAUG----------- .........(((((((((((((.((....)))))))(((((((((..(((....(((......))).....)))..)))))))))..........))-))))))----------- ( -24.40, z-score = -2.26, R) >droWil1.scaffold_181130 6389025 102 + 16660200 -------------CAUAUUAAGCUGAAAUGAUUUAAUGAGCAUGUUUUACCUAAAAUGUAGUAAUUGUCACGUAUUACCUGCUCACCCCGUAUGUAUGUACAUAUAUUGCGUAUU -------------........((.............((((((.((..(((....(((......))).....)))..)).))))))....((((((....))))))...))..... ( -17.10, z-score = -0.26, R) >droGri2.scaffold_14906 10043518 92 - 14172833 ----------CUGAGAAUUAAGCUCAAAUGAUUUAAUGAGCAUGUUU-ACCCAAAAUGUAGUAAUUGUCACGUAUUACGUGCUCAGCCUGAAGAUUU-UGUAUG----------- ----------.((((.......)))).....((((.(((((((((.(-((....(((......))).....)))..)))))))))...)))).....-......----------- ( -16.60, z-score = -0.77, R) >droVir3.scaffold_13047 9117180 93 + 19223366 ----------CAAGGUAUUAAGCUCAAAUGAUUUAAUGAGCAUGUUUUACCUAAAAUGUAGUAAUUGUCACGUAUUACGUGCUCAGCCUGAAGAUUU-UGUACG----------- ----------..((((((((((.((....))))))))((((((((..(((....(((......))).....)))..)))))))).))))........-......----------- ( -20.60, z-score = -1.74, R) >droMoj3.scaffold_6540 2618549 93 - 34148556 ----------CAAGGAAUUAAGCUGAAAUGAUUUAAUGAGCAUGUUUUACCUGAAAUGUAGUAAUUGUCACGUAUUACGUGCUCAGAGUGAAGAUAU-UCUACG----------- ----------...........(..(((....((((.(((((((((..(((.((((((......))).))).)))..)))))))))...))))....)-))..).----------- ( -21.90, z-score = -2.14, R) >consensus _____________CGUAUUAAGCUCAAAUGAUUUAAUGAGCAUGUUUUACCCAAAAUGUAGUAAUUGUCACGUAUUACGUGCUCAGCCUGAAGAUUU_CGUAUG___________ ....................................(((((((((..(((....(((......))).....)))..))))))))).............................. (-14.69 = -14.87 + 0.17)



| Location | 9,318,231 – 9,318,336 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 87.50 |
| Shannon entropy | 0.26552 |
| G+C content | 0.37992 |
| Mean single sequence MFE | -26.46 |
| Consensus MFE | -17.83 |
| Energy contribution | -18.01 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.517866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9318231 105 + 27905053 GAUUUAAUGAGCAUGUUUUACCCAAAAUGUAGUAAUUGUCACGUAUUACGUGCUCAGGCUGAAG------------AUUGCGUAUGCAGAGCUGACAAAUAGCUAAAUCUUUGGGCC .......(((((((((..(((....(((......))).....)))..)))))))))(.(..(((------------(((((....))..(((((.....))))).))))))..).). ( -29.80, z-score = -2.21, R) >droSim1.chr3R 15403867 105 - 27517382 GAUUUAAUGAGCAUGUUUUACCCAAAAUGUAGUAAUUGUCACGUAUUACGUGCUCAGGCUGAAG------------AUUUCGUAUGCAGAGCUGACAAAUAGCUAAAUCUUUGGGCC .......(((((((((..(((....(((......))).....)))..)))))))))(.(..(((------------((((.(....)..(((((.....))))))))))))..).). ( -28.00, z-score = -1.97, R) >droSec1.super_0 8433356 105 - 21120651 GAUUUAAUGAGCAUGUUUUACCCAAAAUGUAGUAAUUGUCACGUAUUACGUGCUCAGGCUGAAG------------AUUUCGUAUGCAGAGCUGACAAAUAGCUAAAUCUUUGGGCC .......(((((((((..(((....(((......))).....)))..)))))))))(.(..(((------------((((.(....)..(((((.....))))))))))))..).). ( -28.00, z-score = -1.97, R) >droYak2.chr3R 13626683 104 + 28832112 GAUUUAAUGAGCAUGUUUUACCCAAA-UGUAGUAAUUGUCACGUAUUACGUGCUCAGGCUGAAG------------AUUGCGUAUGCAGAGCUGACAAAUAGCUAAAUCUUUGGGCC .......(((((((((..(((...((-(......))).....)))..)))))))))(.(..(((------------(((((....))..(((((.....))))).))))))..).). ( -29.40, z-score = -2.06, R) >droEre2.scaffold_4770 5424270 105 - 17746568 GAUUUAAUGAGCAUGUUUUACCCAAAAUGUAGUAAUUGUCACGUAUUACGUGCUCAGGCUGAAG------------AUUGCGUAUGCAGAGCUGACAAAUAGCUAAAUCUUUGGGCC .......(((((((((..(((....(((......))).....)))..)))))))))(.(..(((------------(((((....))..(((((.....))))).))))))..).). ( -29.80, z-score = -2.21, R) >droAna3.scaffold_13340 1875363 107 + 23697760 GAUUUAAUGAGCGUGUUUUACCCAAAAUGUAGUAAUUGUCACGUAUUACGUGCUCAGCCUGAAG----------AUUUUGCUUUGAGAGAGCUGACAAAUAGCUAAAUCUUGGGACU ....................(((((.((.((((..((((((((.....)))((((..(..((((----------......))))..).)))).)))))...)))).)).)))))... ( -24.00, z-score = -0.80, R) >dp4.chr2 16763250 105 + 30794189 GAUUUAAUGAGCAUGUUUUACCUAAAAUGUAGUAAUUGUCACGUAUUACGUGCUCAGCCUGAAG------------AUUGCGUAUGCAGAGCUGACAAAUAGCUAAAUCUGUGGCCU .......(((((((((..(((....(((......))).....)))..)))))))))(((...((------------(((((....))..(((((.....))))).)))))..))).. ( -28.80, z-score = -2.14, R) >droPer1.super_0 8191246 105 - 11822988 GAUUUAAUGAGCAUGUUUUACCUAAAAUGUAGUAAUUGUCACGUAUUACGUGCUCAGCCUGAAG------------AUUGCGUAUGCAGAGCUGACAAAUAGCUAAAUCUGUGGCCU .......(((((((((..(((....(((......))).....)))..)))))))))(((...((------------(((((....))..(((((.....))))).)))))..))).. ( -28.80, z-score = -2.14, R) >droWil1.scaffold_181130 6389041 117 + 16660200 GAUUUAAUGAGCAUGUUUUACCUAAAAUGUAGUAAUUGUCACGUAUUACCUGCUCACCCCGUAUGUAUGUACAUAUAUUGCGUAUUCAGAGCAGACAAAUAUCUAAAUCUUUUGGCC ((((((.((((((.((..(((....(((......))).....)))..)).))))))....((((((....)))))).((((.........)))).........))))))........ ( -20.90, z-score = -0.43, R) >droGri2.scaffold_14906 10043537 99 - 14172833 GAUUUAAUGAGCAUGUUU-ACCCAAAAUGUAGUAAUUGUCACGUAUUACGUGCUCAGCCUGAAG----------AUUUUGUAU--GUAUAGCAGACAAAUAGCUAAAUCUUU----- ((((((.(((((((((.(-((....(((......))).....)))..)))))))))........----------..(((((.(--(.....)).)))))....))))))...----- ( -21.00, z-score = -1.62, R) >droVir3.scaffold_13047 9117199 100 + 19223366 GAUUUAAUGAGCAUGUUUUACCUAAAAUGUAGUAAUUGUCACGUAUUACGUGCUCAGCCUGAAG------------AUUUUGUACGCAUGGCAGACAAAUAGCUAAAUCUCU----- ((((((.(((((((((..(((....(((......))).....)))..)))))))))(((((...------------((...))...)).)))...........))))))...----- ( -23.20, z-score = -1.48, R) >droMoj3.scaffold_6540 2618568 101 - 34148556 GAUUUAAUGAGCAUGUUUUACCUGAAAUGUAGUAAUUGUCACGUAUUACGUGCUCAGAGUGAAG------------AUAUUCUACGCACAGCCGACAAAUAGCUAAAUCUCUA---- ((((((.(((((((((..(((.((((((......))).))).)))..)))))))))..(((.((------------.....)).)))................))))))....---- ( -25.80, z-score = -2.42, R) >consensus GAUUUAAUGAGCAUGUUUUACCCAAAAUGUAGUAAUUGUCACGUAUUACGUGCUCAGCCUGAAG____________AUUGCGUAUGCAGAGCUGACAAAUAGCUAAAUCUUUGGGCC (((((..(((((((((..(((....(((......))).....)))..))))))))).................................(((.........))))))))........ (-17.83 = -18.01 + 0.18)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:11:53 2011