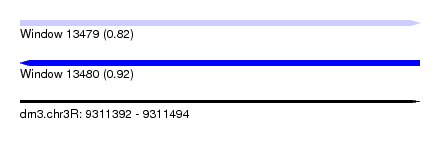
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,311,392 – 9,311,494 |
| Length | 102 |
| Max. P | 0.922873 |

| Location | 9,311,392 – 9,311,494 |
|---|---|
| Length | 102 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 74.14 |
| Shannon entropy | 0.50559 |
| G+C content | 0.44510 |
| Mean single sequence MFE | -23.91 |
| Consensus MFE | -10.60 |
| Energy contribution | -10.74 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.815202 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
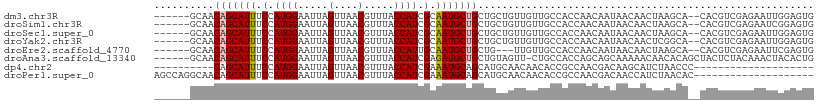
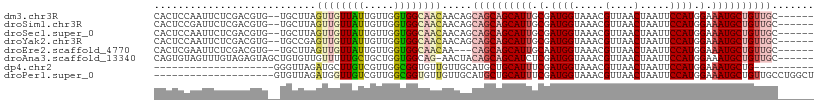
>dm3.chr3R 9311392 102 + 27905053 ------GCAACAGCAUUUCCAUGGAAUUAGUUAACGUUUACCAUCGCAAUGCUGCUGCUGUUGUUGCCACCAACAAUAACAACUAAGCA--CACGUCGAGAAUUGGAGUG ------(((.(((((((.(.((((.....(....).....)))).).))))))).)))((((((((.......))))))))........--(((..(((...)))..))) ( -25.30, z-score = -0.98, R) >droSim1.chr3R 15397039 102 - 27517382 ------GCAACAGCAUUUCCAUGGAAUUAGUUAACGUUUACCAUCGCAAUGCUGCUGCUGUUGUUGCCACCAACAAUAACAACUAAGCA--CACGUCGAGAAUCGGAGUG ------(((.(((((((.(.((((.....(....).....)))).).))))))).)))((((((((.......))))))))........--(((..((.....))..))) ( -27.70, z-score = -1.90, R) >droSec1.super_0 8426761 102 - 21120651 ------GCAACAGCAUUUCCAUGGAAUUAGUUAACGUUUACCAUCGCAAUGCUGCUGCUGUUGUUGCCACCAACAAUAACAACUAAGCA--CACGUCGAGAAUUGGAGUG ------(((.(((((((.(.((((.....(....).....)))).).))))))).)))((((((((.......))))))))........--(((..(((...)))..))) ( -25.30, z-score = -0.98, R) >droYak2.chr3R 13619734 102 + 28832112 ------GCAACAGCAUUUCCAUGGAAUUAGUUAACGUUUACCAUCGCAAUGCUGCUGCUGUUGUUGCCACCAACAAUAACAACUCGGCA--CACGUCGAGAAUUGGAGUG ------(((.(((((((.(.((((.....(....).....)))).).))))))).)))((((((((.......)))))))).((((((.--...)))))).......... ( -31.10, z-score = -2.50, R) >droEre2.scaffold_4770 5417204 99 - 17746568 ------GCAACAGCAUUUCCAUGGAAUUAGUUAACGUUUACCAUUGCAAUGCUGCUG---UUGUUGCCACCAACAAUAACAACUAAGCA--CACGUCGAGAAUUCGAGUG ------((..(((((((.(.((((.....(....).....)))).).))))))).((---((((((.......)))))))).....)).--(((.(((......)))))) ( -25.00, z-score = -1.91, R) >droAna3.scaffold_13340 1869097 103 + 23697760 ------GCAACAGCAUUUCCAUGGAAUUAGUUAACGUUUACCAUCGAGAUGCUGCUGUAGUU-CUGCCACCAGCAGCAAAAACAACACAGCUACUCUACAAACUACACUG ------.....((((((((.((((.....(....).....)))).))))))))(((((.(((-((((.....)))).......))))))))................... ( -24.11, z-score = -2.24, R) >dp4.chr2 16756735 80 + 30794189 ----------CAGCAUUUCCAUGGAAUUAGUUAACGUUUACCAUCGAAAUGCAGCAUGCAACAACACCGCCAACGACAAGCAUCUAACCC-------------------- ----------..(((((((.((((.....(....).....)))).)))))))((.((((....................)))))).....-------------------- ( -15.05, z-score = -2.27, R) >droPer1.super_0 8184293 90 - 11822988 AGCCAGGCAACAGCAUUUCCAUGGAAUUAGUUAACGUUUACCAUCGAAAUGCAGCAUGCAACAACACCGCCAACGACAACCAUCUAACAC-------------------- .....(((....(((((((.((((.....(....).....)))).))))))).(....).........)))...................-------------------- ( -17.70, z-score = -2.30, R) >consensus ______GCAACAGCAUUUCCAUGGAAUUAGUUAACGUUUACCAUCGCAAUGCUGCUGCUGUUGUUGCCACCAACAAUAACAACUAAGCA__CACGUCGAGAAUUGGAGUG ..........(((((((.(.((((.....(....).....)))).).)))))))........................................................ (-10.60 = -10.74 + 0.14)
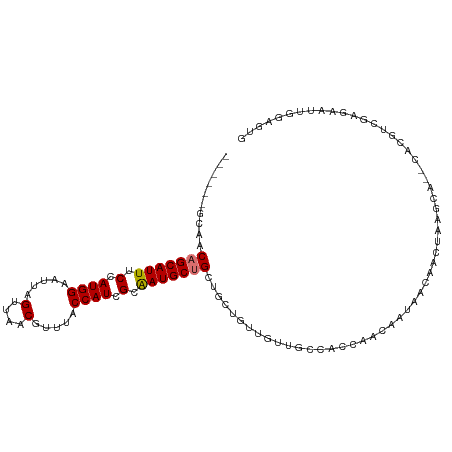
| Location | 9,311,392 – 9,311,494 |
|---|---|
| Length | 102 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 74.14 |
| Shannon entropy | 0.50559 |
| G+C content | 0.44510 |
| Mean single sequence MFE | -28.45 |
| Consensus MFE | -19.52 |
| Energy contribution | -20.40 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.922873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9311392 102 - 27905053 CACUCCAAUUCUCGACGUG--UGCUUAGUUGUUAUUGUUGGUGGCAACAACAGCAGCAGCAUUGCGAUGGUAAACGUUAACUAAUUCCAUGGAAAUGCUGUUGC------ (((((........)).)))--......((((((((.....))))))))....(((((((((((.(.((((.....(....).....)))).).)))))))))))------ ( -29.80, z-score = -1.22, R) >droSim1.chr3R 15397039 102 + 27517382 CACUCCGAUUCUCGACGUG--UGCUUAGUUGUUAUUGUUGGUGGCAACAACAGCAGCAGCAUUGCGAUGGUAAACGUUAACUAAUUCCAUGGAAAUGCUGUUGC------ (((..((.....))..)))--......((((((((.....))))))))....(((((((((((.(.((((.....(....).....)))).).)))))))))))------ ( -31.60, z-score = -1.65, R) >droSec1.super_0 8426761 102 + 21120651 CACUCCAAUUCUCGACGUG--UGCUUAGUUGUUAUUGUUGGUGGCAACAACAGCAGCAGCAUUGCGAUGGUAAACGUUAACUAAUUCCAUGGAAAUGCUGUUGC------ (((((........)).)))--......((((((((.....))))))))....(((((((((((.(.((((.....(....).....)))).).)))))))))))------ ( -29.80, z-score = -1.22, R) >droYak2.chr3R 13619734 102 - 28832112 CACUCCAAUUCUCGACGUG--UGCCGAGUUGUUAUUGUUGGUGGCAACAACAGCAGCAGCAUUGCGAUGGUAAACGUUAACUAAUUCCAUGGAAAUGCUGUUGC------ ...........(((.....--...)))((((((((.....))))))))....(((((((((((.(.((((.....(....).....)))).).)))))))))))------ ( -30.00, z-score = -1.02, R) >droEre2.scaffold_4770 5417204 99 + 17746568 CACUCGAAUUCUCGACGUG--UGCUUAGUUGUUAUUGUUGGUGGCAACAA---CAGCAGCAUUGCAAUGGUAAACGUUAACUAAUUCCAUGGAAAUGCUGUUGC------ (((((((....)))).)))--......((((((((.....))))))))..---((((((((((...((((.....(....).....))))...)))))))))).------ ( -30.40, z-score = -2.16, R) >droAna3.scaffold_13340 1869097 103 - 23697760 CAGUGUAGUUUGUAGAGUAGCUGUGUUGUUUUUGCUGCUGGUGGCAG-AACUACAGCAGCAUCUCGAUGGUAAACGUUAACUAAUUCCAUGGAAAUGCUGUUGC------ (((..(((((........)))))..))).((((((((....))))))-))...(((((((((.((.((((.....(....).....)))).)).))))))))).------ ( -32.60, z-score = -1.74, R) >dp4.chr2 16756735 80 - 30794189 --------------------GGGUUAGAUGCUUGUCGUUGGCGGUGUUGUUGCAUGCUGCAUUUCGAUGGUAAACGUUAACUAAUUCCAUGGAAAUGCUG---------- --------------------.....((((((.(..((....))..).....)))).))(((((((.((((.....(....).....)))).)))))))..---------- ( -19.60, z-score = -0.33, R) >droPer1.super_0 8184293 90 + 11822988 --------------------GUGUUAGAUGGUUGUCGUUGGCGGUGUUGUUGCAUGCUGCAUUUCGAUGGUAAACGUUAACUAAUUCCAUGGAAAUGCUGUUGCCUGGCU --------------------.............((((..((((((((....))))((.(((((((.((((.....(....).....)))).))))))).)))))))))). ( -23.80, z-score = -0.46, R) >consensus CACUCCAAUUCUCGACGUG__UGCUUAGUUGUUAUUGUUGGUGGCAACAACAGCAGCAGCAUUGCGAUGGUAAACGUUAACUAAUUCCAUGGAAAUGCUGUUGC______ ...........................((((((((.....)))))))).....((((((((((.(.((((.....(....).....)))).).))))))))))....... (-19.52 = -20.40 + 0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:11:51 2011