| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,301,920 – 7,302,014 |
| Length | 94 |
| Max. P | 0.905057 |

| Location | 7,301,920 – 7,302,014 |
|---|---|
| Length | 94 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 72.24 |
| Shannon entropy | 0.54789 |
| G+C content | 0.51045 |
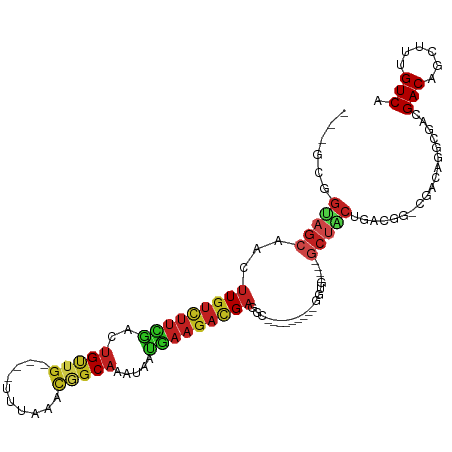
| Mean single sequence MFE | -35.31 |
| Consensus MFE | -11.83 |
| Energy contribution | -11.39 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.57 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.905057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
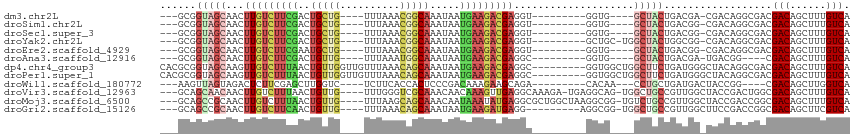
>dm3.chr2L 7301920 94 + 23011544 ---GCGGUAGCAACUUGUCUUCGACUGCUG----UUUAAACGGCAAAUAAUGAAGACGAGGU---------GGUG----GCUACUGACGA-CGACAGGCGACGACAGCUUUGUCA ---.(((((((..((((((((((..(((((----(....)))))).....))))))))))..---------....----)))))))....-.(((((((.......)).))))). ( -36.00, z-score = -3.60, R) >droSim1.chr2L 7097137 94 + 22036055 ---GCGGUAGCAACUUGUCUUCGACUGCUG----UUUAAACGGCAAAUAAUGAAGACGAGGU---------GGUG----GCUACUGACGG-CGACAGGCGACGACAGCUUUGUCA ---.(((((((..((((((((((..(((((----(....)))))).....))))))))))..---------....----)))))))....-.(((((((.......)).))))). ( -36.00, z-score = -3.22, R) >droSec1.super_3 2824297 94 + 7220098 ---GCGGUAGCAACUUGUCUUCGACUGCUG----UUUAAACGGCAAAUAAUGAAGACGAGGU---------GGUG----GCUACUGACGG-CGACAGGCGACGACAGCUUUGUCA ---.(((((((..((((((((((..(((((----(....)))))).....))))))))))..---------....----)))))))....-.(((((((.......)).))))). ( -36.00, z-score = -3.22, R) >droYak2.chr2L 16726103 97 - 22324452 ---GCGGUAGCAACUUGUCUUCGACUGCUG----UUUAAACGGCAAAUAAUGAAGACGAGGU---------GCUGC-UGGCUACUGGCGG-CGACAGGCGACGACAGCUUUGUCA ---.((((((((.((((((((((..(((((----(....)))))).....)))))))))).)---------)))))-))(((......))-)(((((((.......)).))))). ( -41.40, z-score = -3.92, R) >droEre2.scaffold_4929 16222552 94 + 26641161 ---GCGGUAGCAACUUGUCUUCGAAUGCUG----UUUAAACGGCAAAUAAUGAAGACGAGGU---------GGUG----GCUACUGACGG-CGACAGGCGACGACAGCUUUGUCA ---.(((((((..((((((((((..(((((----(....)))))).....))))))))))..---------....----)))))))....-.(((((((.......)).))))). ( -36.00, z-score = -3.45, R) >droAna3.scaffold_12916 15208843 90 + 16180835 ---GCGGUAGCAACUUGUCUUCGACUGUUG----UUUAAAUGGCAAAUAAUGAAGACGAGGC---------GGUG----GCUACUGACGA-UGACGG----CGACAGCUUUGUCA ---.(((((((..((((((((((....(((----((.....)))))....))))))))))..---------....----)))))))..((-..(.((----(....))))..)). ( -32.30, z-score = -3.12, R) >dp4.chr4_group3 10906497 106 - 11692001 CACGCGGUAGCAAGUUGUCUUUAACUGUUGGUUGUUUAAACAGCAAAUAAUGAAGACGAGGC---------GGUGGCUGGCUUCUGAUGGGCUACAGGCGACGACAGCUUUGUCA ..(((.(((((..((((((((((..((((.(((((....))))).)))).)))))))(((((---------........))))).)))..)))))..)))..(((......))). ( -35.30, z-score = -1.84, R) >droPer1.super_1 8020653 106 - 10282868 CACGCGGUAGCAAGUUGUCUUUAACUGUUGGUUGUUUAAACAGCAAAUAAUGAAGACGAGGC---------GGUGGCUGGCUUCUGAUGGGCUACAGGCGACGACAGCUUUGUCA ..(((.(((((..((((((((((..((((.(((((....))))).)))).)))))))(((((---------........))))).)))..)))))..)))..(((......))). ( -35.30, z-score = -1.84, R) >droWil1.scaffold_180772 3480448 92 - 8906247 ---AAGUUAGUAGACUCUUCGAGCUUGGUC----UCUUCACCACUCCCGACAAAGAAGCAGA---------CACAA---CCUGCUGAUGACUACCGG----CGACAGCUUGGUCA ---..((((((((.(((...))).((((((----(((((...............)))).)))---------).)))---.))))))))(((((..((----(....)))))))). ( -23.96, z-score = -0.45, R) >droVir3.scaffold_12963 15578406 106 + 20206255 ---GCAGCAACAACUUGUCUUUAACUGUUG----UUUGGGUCGCAAACAACAAAGUUGAGGCAAAGA-UGAGGCAG-UGGCUGCCGUUGGCUACCGACUGGCGACAGCUUUGUCA ---...((..((((((((.....))(((((----((((.....)))))))))))))))..))...((-..((((.(-(.(((...(((((...))))).))).)).))))..)). ( -36.60, z-score = -1.14, R) >droMoj3.scaffold_6500 18571231 107 + 32352404 ---GCAGCCGCAACUUGUCUUUAACUGUUG----UUUAAGCAGCAAACAAUAAAUAUGAGGCGCUGGCUAAGGCGG-UGUCUGCCGUUGGCUACCGACCGGCGACAGCUUUGUCA ---..((((((..((..(.((((..(((((----(....)))))).....)))).)..))..)).))))(((((..-((((.((((((((...)))).))))))))))))).... ( -34.10, z-score = -0.09, R) >droGri2.scaffold_15126 3527899 98 - 8399593 ---GCAGCCGCAACUUGUCUUCAACUGUUG----UUUAAACAGCAAAUAAUGAAGAUGAGG---------AGGCGG-UGGCUGCCGUUGGCUUCCGACCGGCGACAGCUUCGUCA ---((.(((((..((..((((((..(((((----(....)))))).....))))))..)).---------..))))-).)).((((((((...)))).))))(((......))). ( -40.80, z-score = -3.29, R) >consensus ___GCGGUAGCAACUUGUCUUCGACUGUUG____UUUAAACGGCAAAUAAUGAAGACGAGGC_________GGUG____GCUACUGACGG_CGACAGGCGACGACAGCUUUGUCA ...((....))...(((((((((..(((((..........))))).....)))))))))............((((......)))).................(((......))). (-11.83 = -11.39 + -0.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:23:22 2011