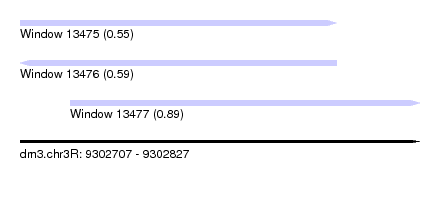
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,302,707 – 9,302,827 |
| Length | 120 |
| Max. P | 0.889378 |

| Location | 9,302,707 – 9,302,802 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.12 |
| Shannon entropy | 0.42035 |
| G+C content | 0.44306 |
| Mean single sequence MFE | -24.31 |
| Consensus MFE | -16.33 |
| Energy contribution | -16.63 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.550492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
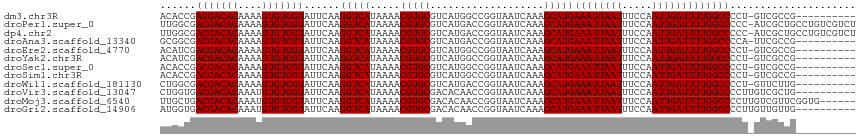
>dm3.chr3R 9302707 95 + 27905053 ------------------AUG-AAGGAGUCCUGGACACCGACGACACAAAAGUGUCGUAUUCAAGGUCAUAAAACGUGCGUCAUGGCCGGUAAUCAAAGCAUGAAAUUAAUUUC ------------------...-.....((..(((..(((.(((((((....)))))))......((((((...((....)).)))))))))..)))..)).............. ( -22.80, z-score = -1.12, R) >droPer1.super_0 8175913 106 - 11822988 -------GGAGCAGGACAGCG-AUGGCGAUGGCGUUGGCGACGACACAAAAGUGUCGUAUUCAAGGUCAUAAAACGUGCGUCAUGACCGGUAAUCAAAGCAUGAAAUUAAUUUC -------...(((.(...((.-...)).(((((.((((..(((((((....)))))))..)))).)))))....).))).(((((....(....)....))))).......... ( -31.00, z-score = -1.88, R) >dp4.chr2 16748596 106 + 30794189 -------GGAGCAGGACAGCG-AUGGCAUUGGCGUUGGCGACGACACAAAAGUGUCGUAUUCAAGGUCAUAAAACGUGCGUCAUGACCGGUAAUCAAAGCAUGAAAUUAAUUUC -------...(((.(...((.-...))..((((.((((..(((((((....)))))))..)))).)))).....).))).(((((....(....)....))))).......... ( -29.60, z-score = -1.67, R) >droAna3.scaffold_13340 1860869 99 + 23697760 --------------GAUCAGG-AGUGAGCUCGGUGCGGCGACGACACAAAAGUGUCGUAUUCAAGGUCAUAAAACGUGCGUCAUGACCGGUAAUCAAAGCAUGAAAUUAAUUUC --------------..((..(-((....))).((((((..(((((((....)))))))..))..((((((...((....)).))))))..........)))))).......... ( -25.70, z-score = -1.19, R) >droEre2.scaffold_4770 5408400 93 - 17746568 --------------------A-AAGGAGUCCUGGACAUCGACGACACAAAAGUGUCGUAUUCAAGGUCAUAAAACGUGCGUCAUGGCCGGUAAUCAAAGCAUGAAAUUAAUUUC --------------------.-...((..((((((.....(((((((....))))))).)))).((((((...((....)).))))))))...))................... ( -20.40, z-score = -1.02, R) >droYak2.chr3R 13610754 93 + 28832112 --------------------A-AAGGAGUCCUGGACAUCGACGACACAAAAGUGUCGUAUUCAAGGUCAUAAAACGUGCGUCAUGGCCGGUAAUCAAAGCAUGAAAUUAAUUUC --------------------.-...((..((((((.....(((((((....))))))).)))).((((((...((....)).))))))))...))................... ( -20.40, z-score = -1.02, R) >droSec1.super_0 8418049 95 - 21120651 ------------------AGG-AAGGAGUCCUGGACACCGACGACACAAAAGUGUCGUAUUCAAGGUCAUAAAACGUGCGUCAUGGCCGGUAAUCAAAGCAUGAAAUUAAUUUC ------------------(((-(.....))))....(((.(((((((....)))))))......((((((...((....)).)))))))))....................... ( -23.00, z-score = -1.19, R) >droSim1.chr3R 15388285 95 - 27517382 ------------------AGG-AAGGAGUCCUGGACACCGACGACACAAAAGUGUCGUAUUCAAGGUCAUAAAACGUGCGUCAUGGCCGGUAAUCAAAGCAUGAAAUUAAUUUC ------------------(((-(.....))))....(((.(((((((....)))))))......((((((...((....)).)))))))))....................... ( -23.00, z-score = -1.19, R) >droWil1.scaffold_181130 6375089 113 + 16660200 AAUGGUAAAUGGGCAAUGGCA-AAGGCAAUGGCACUGGCGACGACACAAAAGUGUCGUAUUCAAGGUCAUAAAACGUGCGUCAUGACCGGUAAUCAAAGCAUGAAAUUAAUUUC ..((((...(((...(((((.-...((((((((..(((..(((((((....)))))))..)))..)))))......))))))))..)))...)))).................. ( -27.80, z-score = -1.46, R) >droVir3.scaffold_13047 9105750 112 + 19223366 --AGACAGCAAGACAACAGGACAACAGGGCAGCGCUGGUGACGACACAAAUGUGUCGUAUUCAAGGUCAUAAAACGUGCGACACAACCGGUAAUCAAAGCAUGAAAUUAAUUUC --.........(((.......((.(((.(...).))).))(((((((....))))))).......)))......(((((...................)))))........... ( -21.61, z-score = 0.16, R) >droGri2.scaffold_14906 10031437 111 - 14172833 ---GACAGGAGGACAAGGGGACAACAGGGCAGCGAUGGUGACGACACAAAUGUGUCGUAUUCAAGGUCAUAAAACGUGCGACACAACCGGUAAUCAAAGCAUGAAAUUAAUUUC ---(((..(((.....(....)..................(((((((....))))))).)))...)))......(((((...................)))))........... ( -22.11, z-score = -0.33, R) >consensus _______________A__GGG_AAGGAGUCCGGGACGGCGACGACACAAAAGUGUCGUAUUCAAGGUCAUAAAACGUGCGUCAUGACCGGUAAUCAAAGCAUGAAAUUAAUUUC ........................................(((((((....))))))).((((.((((((...((....)).)))))).((.......)).))))......... (-16.33 = -16.63 + 0.30)


| Location | 9,302,707 – 9,302,802 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 79.12 |
| Shannon entropy | 0.42035 |
| G+C content | 0.44306 |
| Mean single sequence MFE | -22.05 |
| Consensus MFE | -14.10 |
| Energy contribution | -13.98 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.586542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9302707 95 - 27905053 GAAAUUAAUUUCAUGCUUUGAUUACCGGCCAUGACGCACGUUUUAUGACCUUGAAUACGACACUUUUGUGUCGUCGGUGUCCAGGACUCCUU-CAU------------------ ((((....))))....((((..((((((((((((........)))))...........(((((....))))))))))))..)))).......-...------------------ ( -21.70, z-score = -1.10, R) >droPer1.super_0 8175913 106 + 11822988 GAAAUUAAUUUCAUGCUUUGAUUACCGGUCAUGACGCACGUUUUAUGACCUUGAAUACGACACUUUUGUGUCGUCGCCAACGCCAUCGCCAU-CGCUGUCCUGCUCC------- ((((....))))..((.(((......((((((((........))))))))......(((((((....)))))))...))).)).........-.((......))...------- ( -24.20, z-score = -1.49, R) >dp4.chr2 16748596 106 - 30794189 GAAAUUAAUUUCAUGCUUUGAUUACCGGUCAUGACGCACGUUUUAUGACCUUGAAUACGACACUUUUGUGUCGUCGCCAACGCCAAUGCCAU-CGCUGUCCUGCUCC------- ((((....))))..((.(((......((((((((........))))))))(((...(((((((....)))))))...)))...))).))...-.((......))...------- ( -24.80, z-score = -1.81, R) >droAna3.scaffold_13340 1860869 99 - 23697760 GAAAUUAAUUUCAUGCUUUGAUUACCGGUCAUGACGCACGUUUUAUGACCUUGAAUACGACACUUUUGUGUCGUCGCCGCACCGAGCUCACU-CCUGAUC-------------- ((((....))))..((((.(......((((((((........))))))))......(((((((....))))))).......).)))).....-.......-------------- ( -23.70, z-score = -1.97, R) >droEre2.scaffold_4770 5408400 93 + 17746568 GAAAUUAAUUUCAUGCUUUGAUUACCGGCCAUGACGCACGUUUUAUGACCUUGAAUACGACACUUUUGUGUCGUCGAUGUCCAGGACUCCUU-U-------------------- ..........(((((((.........)).))))).....(((((..(((.((((...((((((....)))))))))).))).))))).....-.-------------------- ( -19.00, z-score = -0.99, R) >droYak2.chr3R 13610754 93 - 28832112 GAAAUUAAUUUCAUGCUUUGAUUACCGGCCAUGACGCACGUUUUAUGACCUUGAAUACGACACUUUUGUGUCGUCGAUGUCCAGGACUCCUU-U-------------------- ..........(((((((.........)).))))).....(((((..(((.((((...((((((....)))))))))).))).))))).....-.-------------------- ( -19.00, z-score = -0.99, R) >droSec1.super_0 8418049 95 + 21120651 GAAAUUAAUUUCAUGCUUUGAUUACCGGCCAUGACGCACGUUUUAUGACCUUGAAUACGACACUUUUGUGUCGUCGGUGUCCAGGACUCCUU-CCU------------------ ((((....))))......((..((((((((((((........)))))...........(((((....))))))))))))..))(((.....)-)).------------------ ( -22.40, z-score = -1.52, R) >droSim1.chr3R 15388285 95 + 27517382 GAAAUUAAUUUCAUGCUUUGAUUACCGGCCAUGACGCACGUUUUAUGACCUUGAAUACGACACUUUUGUGUCGUCGGUGUCCAGGACUCCUU-CCU------------------ ((((....))))......((..((((((((((((........)))))...........(((((....))))))))))))..))(((.....)-)).------------------ ( -22.40, z-score = -1.52, R) >droWil1.scaffold_181130 6375089 113 - 16660200 GAAAUUAAUUUCAUGCUUUGAUUACCGGUCAUGACGCACGUUUUAUGACCUUGAAUACGACACUUUUGUGUCGUCGCCAGUGCCAUUGCCUU-UGCCAUUGCCCAUUUACCAUU ((((....))))..((..........((((((((........))))))))......(((((((....))))))).))(((((.((.......-)).)))))............. ( -24.70, z-score = -2.00, R) >droVir3.scaffold_13047 9105750 112 - 19223366 GAAAUUAAUUUCAUGCUUUGAUUACCGGUUGUGUCGCACGUUUUAUGACCUUGAAUACGACACAUUUGUGUCGUCACCAGCGCUGCCCUGUUGUCCUGUUGUCUUGCUGUCU-- ((((....))))..((...(((.((.((.......(((((((...((((...(....)(((((....)))))))))..)))).)))........)).)).)))..)).....-- ( -21.96, z-score = 0.16, R) >droGri2.scaffold_14906 10031437 111 + 14172833 GAAAUUAAUUUCAUGCUUUGAUUACCGGUUGUGUCGCACGUUUUAUGACCUUGAAUACGACACAUUUGUGUCGUCACCAUCGCUGCCCUGUUGUCCCCUUGUCCUCCUGUC--- ((((....))))..((..((((....(((..((..........))..)))......(((((((....)))))))....))))..)).........................--- ( -18.70, z-score = -0.20, R) >consensus GAAAUUAAUUUCAUGCUUUGAUUACCGGUCAUGACGCACGUUUUAUGACCUUGAAUACGACACUUUUGUGUCGUCGCCGUCCCGGACUCCUU_CCC__U_______________ ((((....))))..............((.(((((........))))).))......(((((((....)))))))........................................ (-14.10 = -13.98 + -0.11)

| Location | 9,302,722 – 9,302,827 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 89.54 |
| Shannon entropy | 0.20682 |
| G+C content | 0.43430 |
| Mean single sequence MFE | -29.60 |
| Consensus MFE | -22.99 |
| Energy contribution | -22.98 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.889378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9302722 105 + 27905053 ACACCGACGACACAAAAGUGUCGUAUUCAAGGUCAUAAAACGUGCGUCAUGGCCGGUAAUCAAAGCAUGAAAUUAAUUUCCAAUUGAUUUUGGCCCCU-GUCGCCG---------- ..(((.(((((((....)))))))......))).......((.(((.((.((((((.((((((.....((((....))))...))))))))))))..)-).)))))---------- ( -31.30, z-score = -2.97, R) >droPer1.super_0 8175939 115 - 11822988 UUGGCGACGACACAAAAGUGUCGUAUUCAAGGUCAUAAAACGUGCGUCAUGACCGGUAAUCAAAGCAUGAAAUUAAUUUCCAAUUGAUUUUGGCCCCC-AUCGCUGCCUGUCGUCU ..(((((((((((....)))))........((((((...((....)).))))))((((......((.((((((((((.....))))))))))))....-.....)))).)))))). ( -30.96, z-score = -1.76, R) >dp4.chr2 16748622 115 + 30794189 UUGGCGACGACACAAAAGUGUCGUAUUCAAGGUCAUAAAACGUGCGUCAUGACCGGUAAUCAAAGCAUGAAAUUAAUUUCCAAUUGAUUUUGGCCCCC-AUCGCUGCCUGUCGUCU ..(((((((((((....)))))........((((((...((....)).))))))((((......((.((((((((((.....))))))))))))....-.....)))).)))))). ( -30.96, z-score = -1.76, R) >droAna3.scaffold_13340 1860888 105 + 23697760 GCGGCGACGACACAAAAGUGUCGUAUUCAAGGUCAUAAAACGUGCGUCAUGACCGGUAAUCAAAGCAUGAAAUUAAUUUCCAAUUGAUUUUGGCCCCA-UUCGCCG---------- .((((((((((((....)))))))......(((((.....(((((....(((.......)))..)))))((((((((.....)))))))))))))...-..)))))---------- ( -32.30, z-score = -3.02, R) >droEre2.scaffold_4770 5408413 105 - 17746568 ACAUCGACGACACAAAAGUGUCGUAUUCAAGGUCAUAAAACGUGCGUCAUGGCCGGUAAUCAAAGCAUGAAAUUAAUUUCCAAUUGAUUUUGGCCCCU-GUCGCCG---------- ......(((((((....)))))))................((.(((.((.((((((.((((((.....((((....))))...))))))))))))..)-).)))))---------- ( -29.70, z-score = -2.55, R) >droYak2.chr3R 13610767 105 + 28832112 ACAUCGACGACACAAAAGUGUCGUAUUCAAGGUCAUAAAACGUGCGUCAUGGCCGGUAAUCAAAGCAUGAAAUUAAUUUCCAAUUGAUUUUGGCCCCU-GUCGCCG---------- ......(((((((....)))))))................((.(((.((.((((((.((((((.....((((....))))...))))))))))))..)-).)))))---------- ( -29.70, z-score = -2.55, R) >droSec1.super_0 8418064 105 - 21120651 ACACCGACGACACAAAAGUGUCGUAUUCAAGGUCAUAAAACGUGCGUCAUGGCCGGUAAUCAAAGCAUGAAAUUAAUUUCCAAUUGAUUUUGGCCCCU-GUCGCCG---------- ..(((.(((((((....)))))))......))).......((.(((.((.((((((.((((((.....((((....))))...))))))))))))..)-).)))))---------- ( -31.30, z-score = -2.97, R) >droSim1.chr3R 15388300 105 - 27517382 ACACCGACGACACAAAAGUGUCGUAUUCAAGGUCAUAAAACGUGCGUCAUGGCCGGUAAUCAAAGCAUGAAAUUAAUUUCCAAUUGAUUUUGGCCCCU-GUCGCCG---------- ..(((.(((((((....)))))))......))).......((.(((.((.((((((.((((((.....((((....))))...))))))))))))..)-).)))))---------- ( -31.30, z-score = -2.97, R) >droWil1.scaffold_181130 6375122 105 + 16660200 CUGGCGACGACACAAAAGUGUCGUAUUCAAGGUCAUAAAACGUGCGUCAUGACCGGUAAUCAAAGCAUGAAAUUAAUUUCCAAUUGAUUUUGGCCCCU-GUUCUUG---------- ..(((.(((((((....)))))))......((((((...((....)).))))))..............(((((((((.....))))))))).)))...-.......---------- ( -25.30, z-score = -1.26, R) >droVir3.scaffold_13047 9105782 106 + 19223366 CUGGUGACGACACAAAUGUGUCGUAUUCAAGGUCAUAAAACGUGCGACACAACCGGUAAUCAAAGCAUGAAAUUAAUUUCCAAUUGAUUUUGGCCCCUUGUCGCUG---------- ..(((((((((((....)))))........(((((.....(((((...................)))))((((((((.....)))))))))))))....)))))).---------- ( -28.11, z-score = -1.81, R) >droMoj3.scaffold_6540 2605937 110 - 34148556 UUGCUGACGACACAAAUGUGUCGUAUUCAAGGUCAUAAAACGUGCGACACAACCGGUAAUCAAAGCAUGAAAUUAAUUUCCAAUUGAUUUUGGCCCCUUGUCGUUCGGUG------ ..((((((((((....((((((((((...............)))))))))).((((.((((((.....((((....))))...)))))))))).....))))).))))).------ ( -29.86, z-score = -1.80, R) >droGri2.scaffold_14906 10031468 106 - 14172833 AUGGUGACGACACAAAUGUGUCGUAUUCAAGGUCAUAAAACGUGCGACACAACCGGUAAUCAAAGCAUGAAAUUAAUUUCCAAUUGAUUUUGGCCCCUUGUUGUUG---------- .....(((((((....((((((((((...............)))))))))).((((.((((((.....((((....))))...)))))))))).....))))))).---------- ( -24.46, z-score = -0.46, R) >consensus ACGGCGACGACACAAAAGUGUCGUAUUCAAGGUCAUAAAACGUGCGUCAUGACCGGUAAUCAAAGCAUGAAAUUAAUUUCCAAUUGAUUUUGGCCCCU_GUCGCCG__________ ......(((((((....)))))))......(((((.....(((((...................)))))((((((((.....)))))))))))))..................... (-22.99 = -22.98 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:11:48 2011