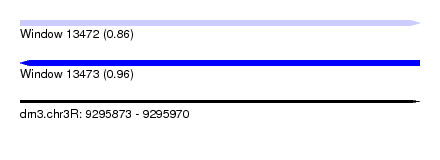
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,295,873 – 9,295,970 |
| Length | 97 |
| Max. P | 0.956602 |

| Location | 9,295,873 – 9,295,970 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 81.72 |
| Shannon entropy | 0.31007 |
| G+C content | 0.49352 |
| Mean single sequence MFE | -22.24 |
| Consensus MFE | -14.72 |
| Energy contribution | -16.12 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.863508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
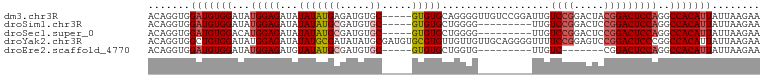
>dm3.chr3R 9295873 97 + 27905053 UUCUUAAUAAUGUGGCCUGGAGUCCGUAGUCCGGACAAUCCGGACAACCCCUGCACAC-----GCACAUCUCAUAUAUAUCUCCAUAUCCACAUCCACCUGU .........((((((..(((((...(((((((((.....))))))......(((....-----))).........)))..)))))...))))))........ ( -27.50, z-score = -3.78, R) >droSim1.chr3R 15381440 88 - 27517382 UUCUUAAUAAUGUGGCCUGGAGUCCGGAGUCCGGACAA---------CCCCAGCACAC-----GCACAUCGCAUAUAUAUCUCCAUAUCCACAUCCACCUGU .........((((((..((((((((((...))))))..---------...........-----((.....)).........))))...))))))........ ( -22.40, z-score = -2.12, R) >droSec1.super_0 8411256 88 - 21120651 UUCUUAAUAAUGUGGCCUGGAGUCCGGAGUCCGGACAA---------CCCCAGCACAC-----GCACAUCGCAUAUAUAUCUCCAUGUCCACAUCCACCUGU .........(((((((.((((((((((...))))))..---------...........-----((.....)).........)))).).))))))........ ( -23.90, z-score = -2.13, R) >droYak2.chr3R 13603988 102 + 28832112 UUCUUAAUAAUGUGGCCGGGAGUCCGGACUCCGGAAAACCCCUGCAACAACAACACGCACAUCGCAUAUAUCGCAUAUAUCUCCAUAUCCACAGCCACCUGU .........(((((((.(((..(((((...)))))....))).))...........((.....))......)))))..............((((....)))) ( -21.40, z-score = -0.88, R) >droEre2.scaffold_4770 5401678 81 - 17746568 UUCUUAAUAAUGUGGCCUGGAGUCCG-------GACAA---------CACCAGCACAC-----GCACAUCGCAUAUACAUCUCCAUAUCCACAUCCACCUGU .........((((((..(((((...(-------(....---------..)).......-----((.....))........)))))...))))))........ ( -16.00, z-score = -1.53, R) >consensus UUCUUAAUAAUGUGGCCUGGAGUCCGGAGUCCGGACAA_________CCCCAGCACAC_____GCACAUCGCAUAUAUAUCUCCAUAUCCACAUCCACCUGU .........((((((..(((((((((.....)))))...........................((.....)).........))))...))))))........ (-14.72 = -16.12 + 1.40)

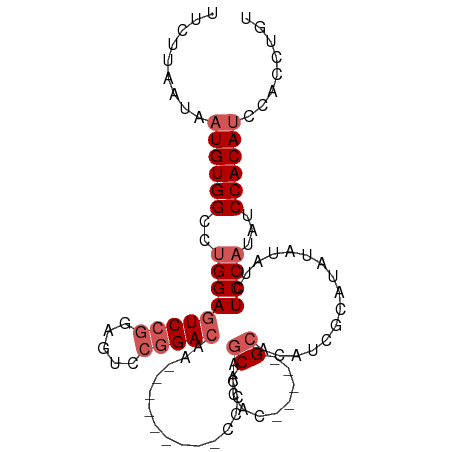
| Location | 9,295,873 – 9,295,970 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 81.72 |
| Shannon entropy | 0.31007 |
| G+C content | 0.49352 |
| Mean single sequence MFE | -33.26 |
| Consensus MFE | -21.30 |
| Energy contribution | -21.78 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.956602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9295873 97 - 27905053 ACAGGUGGAUGUGGAUAUGGAGAUAUAUAUGAGAUGUGC-----GUGUGCAGGGGUUGUCCGGAUUGUCCGGACUACGGACUCCAGGCCACAUUAUUAAGAA .......(((((((...(((((............(.(((-----....))).).((.((((((.....)))))).))...)))))..)))))))........ ( -33.70, z-score = -2.47, R) >droSim1.chr3R 15381440 88 + 27517382 ACAGGUGGAUGUGGAUAUGGAGAUAUAUAUGCGAUGUGC-----GUGUGCUGGGG---------UUGUCCGGACUCCGGACUCCAGGCCACAUUAUUAAGAA .......(((((((...(((((...(((((((.....))-----)))))((((((---------((.....)))))))).)))))..)))))))........ ( -34.10, z-score = -2.96, R) >droSec1.super_0 8411256 88 + 21120651 ACAGGUGGAUGUGGACAUGGAGAUAUAUAUGCGAUGUGC-----GUGUGCUGGGG---------UUGUCCGGACUCCGGACUCCAGGCCACAUUAUUAAGAA .......(((((((...(((((...(((((((.....))-----)))))((((((---------((.....)))))))).)))))..)))))))........ ( -34.10, z-score = -2.72, R) >droYak2.chr3R 13603988 102 - 28832112 ACAGGUGGCUGUGGAUAUGGAGAUAUAUGCGAUAUAUGCGAUGUGCGUGUUGUUGUUGCAGGGGUUUUCCGGAGUCCGGACUCCCGGCCACAUUAUUAAGAA ....(((((((.......(((((....(((((((.(((((.....)))))...))))))).....)))))(((((....))))))))))))........... ( -35.60, z-score = -1.96, R) >droEre2.scaffold_4770 5401678 81 + 17746568 ACAGGUGGAUGUGGAUAUGGAGAUGUAUAUGCGAUGUGC-----GUGUGCUGGUG---------UUGUC-------CGGACUCCAGGCCACAUUAUUAAGAA .......(((((((...(((((..((((((((.....))-----))))))(((..---------....)-------))..)))))..)))))))........ ( -28.80, z-score = -2.54, R) >consensus ACAGGUGGAUGUGGAUAUGGAGAUAUAUAUGCGAUGUGC_____GUGUGCUGGGG_________UUGUCCGGACUCCGGACUCCAGGCCACAUUAUUAAGAA .......(((((((...(((((.............(..(.......)..).................((((.....)))))))))..)))))))........ (-21.30 = -21.78 + 0.48)
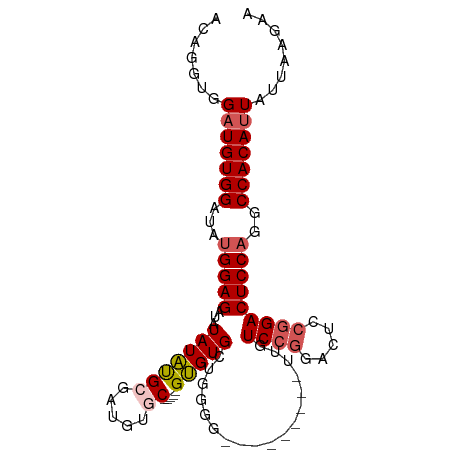
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:11:45 2011