
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,292,580 – 9,292,638 |
| Length | 58 |
| Max. P | 0.725724 |

| Location | 9,292,580 – 9,292,638 |
|---|---|
| Length | 58 |
| Sequences | 5 |
| Columns | 61 |
| Reading direction | forward |
| Mean pairwise identity | 68.80 |
| Shannon entropy | 0.51975 |
| G+C content | 0.38844 |
| Mean single sequence MFE | -7.86 |
| Consensus MFE | -3.78 |
| Energy contribution | -3.06 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 2.00 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.725724 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
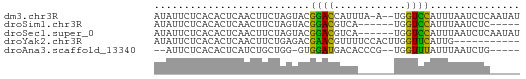
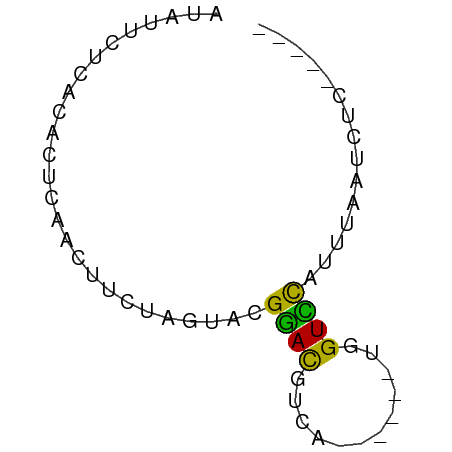
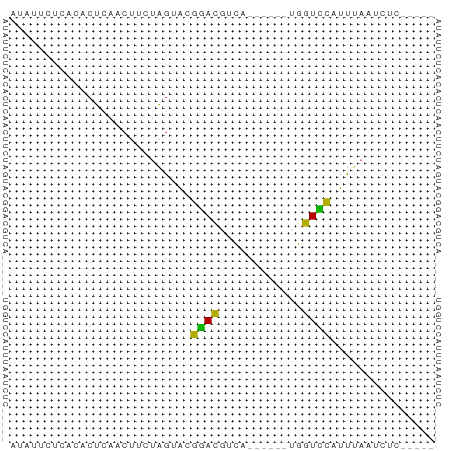
>dm3.chr3R 9292580 58 + 27905053 AUAUUCUCACACUCAACUUCUAGUACGGACCAUUUA-A--UGGUCCAUUUAAUCUCAAUAU ..........................(((((((...-)--))))))............... ( -10.20, z-score = -3.77, R) >droSim1.chr3R 15378174 50 - 27517382 AUAUUCUCACACUCAACUUCUAGUACGGACGUCA------UGGUCCAUUUAAUCUC----- ..........................((((....------..))))..........----- ( -6.10, z-score = -1.17, R) >droSec1.super_0 8407972 55 - 21120651 AUAUUCUCACACUCAACUUCUAGUACGGACGUCA------UGGUCCAUUUAAUCUCAAUAU ..........................((((....------..))))............... ( -6.10, z-score = -1.15, R) >droYak2.chr3R 13600728 50 + 28832112 AUAUUCUCACACUCAACUUCUGAGACGAACGUUUUCCACUUGGUUCAUUG----------- ...........((((.....))))..((((............))))....----------- ( -5.50, z-score = -0.27, R) >droAna3.scaffold_13340 1851199 51 + 23697760 --AUUCUCACACUCAUCUGCUGG-GUGGAUGACACCCG--UGGUUUAUUUAAUCUG----- --..............(..(.((-(((.....))))))--..).............----- ( -11.40, z-score = -1.14, R) >consensus AUAUUCUCACACUCAACUUCUAGUACGGACGUCA______UGGUCCAUUUAAUCUC_____ ..........................((((............))))............... ( -3.78 = -3.06 + -0.72)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:11:44 2011