
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,289,944 – 9,290,026 |
| Length | 82 |
| Max. P | 0.999264 |

| Location | 9,289,944 – 9,290,026 |
|---|---|
| Length | 82 |
| Sequences | 14 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 88.48 |
| Shannon entropy | 0.26611 |
| G+C content | 0.50874 |
| Mean single sequence MFE | -30.51 |
| Consensus MFE | -25.02 |
| Energy contribution | -25.48 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.45 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.27 |
| SVM RNA-class probability | 0.998155 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
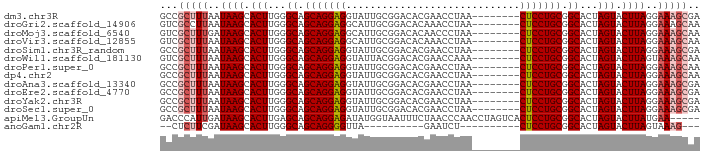
>dm3.chr3R 9289944 82 + 27905053 GCCGCUUUAAUAAGCACUUGGGCAGCAGGAGGUAUUGCGGACACGAACCUAA--------CUCCUGCGGCACUAGUACUUAGGAAAGCGA ..((((((..((((.(((...((.(((((((......((....)).......--------))))))).))...))).))))..)))))). ( -33.72, z-score = -4.06, R) >droGri2.scaffold_14906 3807837 82 + 14172833 GUCGCCUUAAUAAGCACUUGGGCAGCAGGAGGCAUUGCGGACACAAACCUAA--------CUCCUGCGGCACUAGUACUUAGGAAAGCAA ...((.((..((((.(((...((.(((((((((...))((.......))...--------))))))).))...))).))))..)).)).. ( -26.30, z-score = -1.79, R) >droMoj3.scaffold_6540 10349278 82 + 34148556 GUCGCUUUGAUAAGCACUUGGGCAGCAGGAGGCAUUGCGGACACAACCCUAA--------CUCCUGCGGCACUAGUACUUAGGAAAGCAA ...(((((..((((.(((...((.(((((((.....(.((......)))...--------))))))).))...))).))))..))))).. ( -30.40, z-score = -2.82, R) >droVir3.scaffold_12855 7299611 82 - 10161210 GUCGCUUUAAUAAGCACUUGGGCAGCAGGAGGCAUUGCGGACACAAACCUAA--------CUCCUGCGGCACUAGUACUUAGGAAAGCAA ...(((((..((((.(((...((.(((((((((...))((.......))...--------))))))).))...))).))))..))))).. ( -30.40, z-score = -3.56, R) >droSim1.chr3R_random 533445 82 + 1307089 GCCGCUUUAAUAAGCACUUGGGCAGCAGGAGGUAUUGCGGACACGAACCUAA--------CUCCUGCGGCACUAGUACUUAGGAAAGCGA ..((((((..((((.(((...((.(((((((......((....)).......--------))))))).))...))).))))..)))))). ( -33.72, z-score = -4.06, R) >droWil1.scaffold_181130 6363229 82 + 16660200 GUCGCUUUAAUAAGCACUUGGGCAGCAGGAGGUAUUACGGACACGAACCAAA--------CUCCUGCGGCACUAGUACUUAGUAAAGCAA ...((((((.((((.(((...((.(((((((......((....)).......--------))))))).))...))).)))).)))))).. ( -30.92, z-score = -4.51, R) >droPer1.super_0 8163706 82 - 11822988 GCCGCUUUAAUAAGCACUUGGGCAGCAGGAGGUAUUGCGGACACGAACCUAA--------CUCCUGCGGCACUAGUACUUAGGAAAGCAA ...(((((..((((.(((...((.(((((((......((....)).......--------))))))).))...))).))))..))))).. ( -31.92, z-score = -3.61, R) >dp4.chr2 16736635 82 + 30794189 GCCGCUUUAAUAAGCACUUGGGCAGCAGGAGGUAUUGCGGACACGAACCUAA--------CUCCUGCGGCACUAGUACUUAGGAAAGCAA ...(((((..((((.(((...((.(((((((......((....)).......--------))))))).))...))).))))..))))).. ( -31.92, z-score = -3.61, R) >droAna3.scaffold_13340 1848681 82 + 23697760 GCCGCUUUAAUAAGCACUUGGGCAGCAGGAGGUAUUGCGGACACGAACCUAA--------CUCCUGCGGCACUAGUACUUAGGAAAGCGA ..((((((..((((.(((...((.(((((((......((....)).......--------))))))).))...))).))))..)))))). ( -33.72, z-score = -4.06, R) >droEre2.scaffold_4770 5395799 82 - 17746568 GCCGCUUUAAUAAGCACUUGGGCAGCAGGAGGUAUUGCGGACACGAACCUAA--------CUCCUGCGGCACUAGUACUUAGGAAAGCGA ..((((((..((((.(((...((.(((((((......((....)).......--------))))))).))...))).))))..)))))). ( -33.72, z-score = -4.06, R) >droYak2.chr3R 13598024 82 + 28832112 GCCGCUUUAAUAAGCACUUGGGCAGCAGGAGGUAUUGCGGACACGAACCUAA--------CUCCUGCGGCACUAGUACUUAGGAAAGCGA ..((((((..((((.(((...((.(((((((......((....)).......--------))))))).))...))).))))..)))))). ( -33.72, z-score = -4.06, R) >droSec1.super_0 8405346 82 - 21120651 GCCGCUUUAAUAAGCACUUGGGCAGCAGGAGGUAUUGCGGACACGAACCUAA--------CUCCUGCGGCACUAGUACUUAGGAAAGCGA ..((((((..((((.(((...((.(((((((......((....)).......--------))))))).))...))).))))..)))))). ( -33.72, z-score = -4.06, R) >apiMel3.GroupUn 149494248 85 + 399230636 GACCCAUUGAUAAGCACUUGAGCAGCAGGAGAUAUGGUAAUUUCUAACCCAACCUAGUCACUCCUGCGGCACUAGUACUUAUGAA----- ......((.(((((.(((.(.((.(((((((....(((.............)))......))))))).)).).))).))))).))----- ( -23.02, z-score = -2.26, R) >anoGam1.chr2R 54489126 65 + 62725911 --CUCUUCGAUAAGCACUUGGGCAGCAGGGGUUA----------GAAUCU----------CUCCUGCGGCACUAGUACUUAGUAAAG--- --..(((...((((.(((...((.(((((((...----------......----------))))))).))...))).))))...)))--- ( -20.00, z-score = -1.79, R) >consensus GCCGCUUUAAUAAGCACUUGGGCAGCAGGAGGUAUUGCGGACACGAACCUAA________CUCCUGCGGCACUAGUACUUAGGAAAGCAA ...(((((..((((.(((...((.(((((((((...))((.......))...........))))))).))...))).))))..))))).. (-25.02 = -25.48 + 0.46)

| Location | 9,289,944 – 9,290,026 |
|---|---|
| Length | 82 |
| Sequences | 14 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 88.48 |
| Shannon entropy | 0.26611 |
| G+C content | 0.50874 |
| Mean single sequence MFE | -32.68 |
| Consensus MFE | -29.53 |
| Energy contribution | -29.67 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.22 |
| Mean z-score | -4.51 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.75 |
| SVM RNA-class probability | 0.999264 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
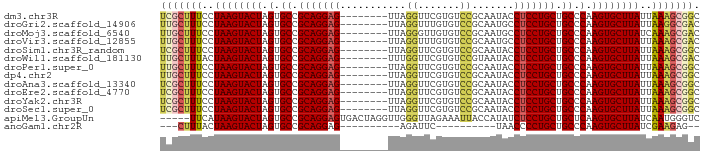
>dm3.chr3R 9289944 82 - 27905053 UCGCUUUCCUAAGUACUAGUGCCGCAGGAG--------UUAGGUUCGUGUCCGCAAUACCUCCUGCUGCCCAAGUGCUUAUUAAAGCGGC (((((((..((((((((.(.((.(((((((--------....(((((....)).)))..))))))).)).).))))))))..))))))). ( -34.00, z-score = -4.75, R) >droGri2.scaffold_14906 3807837 82 - 14172833 UUGCUUUCCUAAGUACUAGUGCCGCAGGAG--------UUAGGUUUGUGUCCGCAAUGCCUCCUGCUGCCCAAGUGCUUAUUAAGGCGAC (((((((..((((((((.(.((.((((((.--------...(((((((....)))).))))))))).)).).))))))))..))))))). ( -33.90, z-score = -4.31, R) >droMoj3.scaffold_6540 10349278 82 - 34148556 UUGCUUUCCUAAGUACUAGUGCCGCAGGAG--------UUAGGGUUGUGUCCGCAAUGCCUCCUGCUGCCCAAGUGCUUAUCAAAGCGAC (((((((..((((((((.(.((.(((((((--------...(.(((((....))))).)))))))).)).).))))))))..))))))). ( -36.70, z-score = -5.21, R) >droVir3.scaffold_12855 7299611 82 + 10161210 UUGCUUUCCUAAGUACUAGUGCCGCAGGAG--------UUAGGUUUGUGUCCGCAAUGCCUCCUGCUGCCCAAGUGCUUAUUAAAGCGAC (((((((..((((((((.(.((.((((((.--------...(((((((....)))).))))))))).)).).))))))))..))))))). ( -34.20, z-score = -5.12, R) >droSim1.chr3R_random 533445 82 - 1307089 UCGCUUUCCUAAGUACUAGUGCCGCAGGAG--------UUAGGUUCGUGUCCGCAAUACCUCCUGCUGCCCAAGUGCUUAUUAAAGCGGC (((((((..((((((((.(.((.(((((((--------....(((((....)).)))..))))))).)).).))))))))..))))))). ( -34.00, z-score = -4.75, R) >droWil1.scaffold_181130 6363229 82 - 16660200 UUGCUUUACUAAGUACUAGUGCCGCAGGAG--------UUUGGUUCGUGUCCGUAAUACCUCCUGCUGCCCAAGUGCUUAUUAAAGCGAC ((((((((.((((((((.(.((.(((((((--------....(((((....)).)))..))))))).)).).)))))))).)))))))). ( -34.60, z-score = -5.81, R) >droPer1.super_0 8163706 82 + 11822988 UUGCUUUCCUAAGUACUAGUGCCGCAGGAG--------UUAGGUUCGUGUCCGCAAUACCUCCUGCUGCCCAAGUGCUUAUUAAAGCGGC .((((((..((((((((.(.((.(((((((--------....(((((....)).)))..))))))).)).).))))))))..)))))).. ( -32.00, z-score = -3.96, R) >dp4.chr2 16736635 82 - 30794189 UUGCUUUCCUAAGUACUAGUGCCGCAGGAG--------UUAGGUUCGUGUCCGCAAUACCUCCUGCUGCCCAAGUGCUUAUUAAAGCGGC .((((((..((((((((.(.((.(((((((--------....(((((....)).)))..))))))).)).).))))))))..)))))).. ( -32.00, z-score = -3.96, R) >droAna3.scaffold_13340 1848681 82 - 23697760 UCGCUUUCCUAAGUACUAGUGCCGCAGGAG--------UUAGGUUCGUGUCCGCAAUACCUCCUGCUGCCCAAGUGCUUAUUAAAGCGGC (((((((..((((((((.(.((.(((((((--------....(((((....)).)))..))))))).)).).))))))))..))))))). ( -34.00, z-score = -4.75, R) >droEre2.scaffold_4770 5395799 82 + 17746568 UCGCUUUCCUAAGUACUAGUGCCGCAGGAG--------UUAGGUUCGUGUCCGCAAUACCUCCUGCUGCCCAAGUGCUUAUUAAAGCGGC (((((((..((((((((.(.((.(((((((--------....(((((....)).)))..))))))).)).).))))))))..))))))). ( -34.00, z-score = -4.75, R) >droYak2.chr3R 13598024 82 - 28832112 UCGCUUUCCUAAGUACUAGUGCCGCAGGAG--------UUAGGUUCGUGUCCGCAAUACCUCCUGCUGCCCAAGUGCUUAUUAAAGCGGC (((((((..((((((((.(.((.(((((((--------....(((((....)).)))..))))))).)).).))))))))..))))))). ( -34.00, z-score = -4.75, R) >droSec1.super_0 8405346 82 + 21120651 UCGCUUUCCUAAGUACUAGUGCCGCAGGAG--------UUAGGUUCGUGUCCGCAAUACCUCCUGCUGCCCAAGUGCUUAUUAAAGCGGC (((((((..((((((((.(.((.(((((((--------....(((((....)).)))..))))))).)).).))))))))..))))))). ( -34.00, z-score = -4.75, R) >apiMel3.GroupUn 149494248 85 - 399230636 -----UUCAUAAGUACUAGUGCCGCAGGAGUGACUAGGUUGGGUUAGAAAUUACCAUAUCUCCUGCUGCUCAAGUGCUUAUCAAUGGGUC -----...(((((((((.(.((.(((((((...(......)(((.(....).)))....))))))).)).).)))))))))......... ( -28.10, z-score = -2.45, R) >anoGam1.chr2R 54489126 65 - 62725911 ---CUUUACUAAGUACUAGUGCCGCAGGAG----------AGAUUC----------UAACCCCUGCUGCCCAAGUGCUUAUCGAAGAG-- ---((((..((((((((.(.((.(((((..----------......----------.....))))).)).).))))))))..))))..-- ( -22.02, z-score = -3.83, R) >consensus UCGCUUUCCUAAGUACUAGUGCCGCAGGAG________UUAGGUUCGUGUCCGCAAUACCUCCUGCUGCCCAAGUGCUUAUUAAAGCGGC (((((((..((((((((.(.((.(((((((...........((.......)).......))))))).)).).))))))))..))))))). (-29.53 = -29.67 + 0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:11:43 2011