
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,289,530 – 9,289,665 |
| Length | 135 |
| Max. P | 0.826114 |

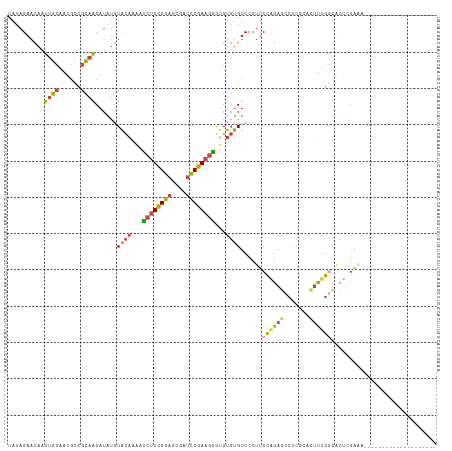
| Location | 9,289,530 – 9,289,628 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 67.90 |
| Shannon entropy | 0.55718 |
| G+C content | 0.45479 |
| Mean single sequence MFE | -22.98 |
| Consensus MFE | -15.54 |
| Energy contribution | -15.30 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.50 |
| Mean z-score | -0.74 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.826114 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9289530 98 + 27905053 UAUAGAACAAUUGCAAUGCCGCAACAUAUGUACAAAAUCUUCGGAGCGAUCCGAAGGGUGUGUGUCCCUUCCAGAGCCUCGCACUUUGGCACUUGAAA-------------------- ..........((((......))))...(..((((...((((((((....)))))))).))))..)...(((.((.(((.........))).)).))).-------------------- ( -25.20, z-score = -0.83, R) >droAna3.scaffold_13340 1848315 90 + 23697760 UAUAGAACAAUUGCUAGGCCGCAACAUAUGUACAAU-UCUUCGGAGCGAUCCGAAGGGAGUGUGUCCCUUCCAGAAACCCCGAACUGGAAG--------------------------- ................((.((((((....))....(-((((((((....)))))))))..)))).))(((((((..........)))))))--------------------------- ( -24.10, z-score = -1.53, R) >droEre2.scaffold_4770 5395381 98 - 17746568 UAUAGAACAAUUGCAAUGCCGCAACAUAUGUACAAAAUCUUCGGAGCGAUCCGAAGGAUGUGUGUCCCUUCCAGAGCCUCGCACUUUGGCACUCGAAA-------------------- ..........((((......))))......((((...((((((((....))))))))...))))....(((.((.(((.........))).)).))).-------------------- ( -23.20, z-score = -0.65, R) >droYak2.chr3R 13597608 98 + 28832112 UAUAGAACAAUUGCAAUGCCGCAACAUAUGUACAAAAUCUUCGGAGCGAUCCGAAGAGUGUGUGUCCCUUCCAGAGCCUCGCACUUUGGCACUCGAAA-------------------- ..........((((......))))...(..((((...((((((((....)))))))).))))..)...(((.((.(((.........))).)).))).-------------------- ( -26.10, z-score = -1.73, R) >droSec1.super_0 8404934 98 - 21120651 UAUAGAACAAUUGCAAUGCCGCAACAUAUGUACAAAAUCUUCGGAGCGAUCCGAAGGGUGUGUGUCCCUUCCAGAGCCUCGCACUUUGGCACUCGAAG-------------------- ..........((((......))))...(..((((...((((((((....)))))))).))))..)..((((.((.(((.........))).)).))))-------------------- ( -27.20, z-score = -1.47, R) >droGri2.scaffold_14906 3807330 117 + 14172833 UA-AGAUUAAUAUUUACAUGUACGCAAUGAUUUAGAACAAUUGAAGCCACUCAAAAGAAGGCAGUCCCCACUUAAAAACAAUGUUUGGCUAACUGUGUCCUCAUCCAAAAACUUGUAA ..-..........(((((.(((((((........(((((.(((..(((..((....)).)))(((....)))......)))))))).......)))))............)).))))) ( -12.06, z-score = 1.79, R) >consensus UAUAGAACAAUUGCAAUGCCGCAACAUAUGUACAAAAUCUUCGGAGCGAUCCGAAGGGUGUGUGUCCCUUCCAGAGCCUCGCACUUUGGCACUCGAAA____________________ ..........((((......))))......((((...((((((((....))))))))...))))......((((((.......))))))............................. (-15.54 = -15.30 + -0.24)

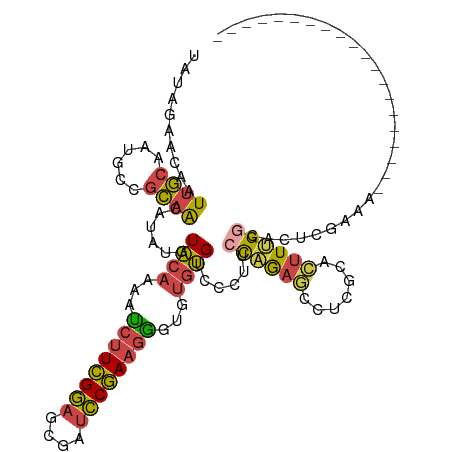
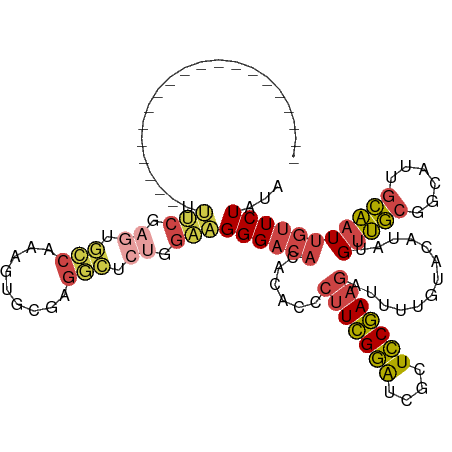
| Location | 9,289,530 – 9,289,628 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 67.90 |
| Shannon entropy | 0.55718 |
| G+C content | 0.45479 |
| Mean single sequence MFE | -26.98 |
| Consensus MFE | -17.67 |
| Energy contribution | -18.37 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.35 |
| Mean z-score | -0.79 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.807531 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9289530 98 - 27905053 --------------------UUUCAAGUGCCAAAGUGCGAGGCUCUGGAAGGGACACACACCCUUCGGAUCGCUCCGAAGAUUUUGUACAUAUGUUGCGGCAUUGCAAUUGUUCUAUA --------------------.....((((((...((((((((.(((.((((((.......))))))(((....)))..)))))))))))(.....)..)))))).............. ( -25.40, z-score = -0.21, R) >droAna3.scaffold_13340 1848315 90 - 23697760 ---------------------------CUUCCAGUUCGGGGUUUCUGGAAGGGACACACUCCCUUCGGAUCGCUCCGAAGA-AUUGUACAUAUGUUGCGGCCUAGCAAUUGUUCUAUA ---------------------------........(((((((.((((.((((((.....))))))))))..)))))))(((-((.........(((((......))))).)))))... ( -29.00, z-score = -1.96, R) >droEre2.scaffold_4770 5395381 98 + 17746568 --------------------UUUCGAGUGCCAAAGUGCGAGGCUCUGGAAGGGACACACAUCCUUCGGAUCGCUCCGAAGAUUUUGUACAUAUGUUGCGGCAUUGCAAUUGUUCUAUA --------------------.(((.((.(((.........))).)).)))((((((.(((..(((((((....)))))))....)))......((((((....))))))))))))... ( -27.70, z-score = -0.75, R) >droYak2.chr3R 13597608 98 - 28832112 --------------------UUUCGAGUGCCAAAGUGCGAGGCUCUGGAAGGGACACACACUCUUCGGAUCGCUCCGAAGAUUUUGUACAUAUGUUGCGGCAUUGCAAUUGUUCUAUA --------------------.(((.((.(((.........))).)).)))((((((((.(.((((((((....)))))))).).)))......((((((....)))))).)))))... ( -29.10, z-score = -1.28, R) >droSec1.super_0 8404934 98 + 21120651 --------------------CUUCGAGUGCCAAAGUGCGAGGCUCUGGAAGGGACACACACCCUUCGGAUCGCUCCGAAGAUUUUGUACAUAUGUUGCGGCAUUGCAAUUGUUCUAUA --------------------((((.((.(((.........))).)).))))(((((......(((((((....))))))).............((((((....))))))))))).... ( -28.80, z-score = -0.98, R) >droGri2.scaffold_14906 3807330 117 - 14172833 UUACAAGUUUUUGGAUGAGGACACAGUUAGCCAAACAUUGUUUUUAAGUGGGGACUGCCUUCUUUUGAGUGGCUUCAAUUGUUCUAAAUCAUUGCGUACAUGUAAAUAUUAAUCU-UA (((((.((.....(((..(((.(((((((((((..((........(((.(((.....))).))).))..)))))..)))))))))..))).......)).)))))..........-.. ( -21.90, z-score = 0.41, R) >consensus ____________________UUUCGAGUGCCAAAGUGCGAGGCUCUGGAAGGGACACACACCCUUCGGAUCGCUCCGAAGAUUUUGUACAUAUGUUGCGGCAUUGCAAUUGUUCUAUA .....................(((.((.(((.........))).)).)))((((((......(((((((....))))))).............(((((......)))))))))))... (-17.67 = -18.37 + 0.70)


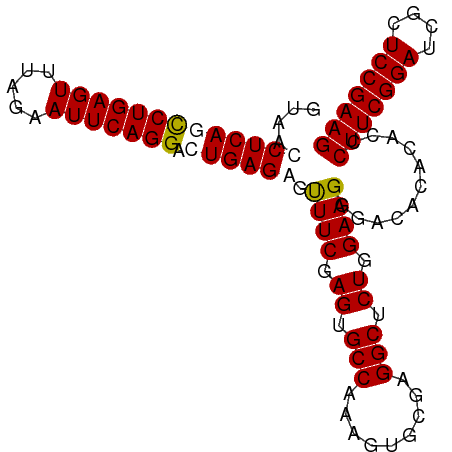
| Location | 9,289,568 – 9,289,665 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 95.19 |
| Shannon entropy | 0.07790 |
| G+C content | 0.53608 |
| Mean single sequence MFE | -34.35 |
| Consensus MFE | -31.30 |
| Energy contribution | -31.92 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.745378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9289568 97 - 27905053 GUAACCUCAGUCUGAGUUUAGAAUUCAGGACUGAGGGUUUCAAGUGCCAAAGUGCGAGGCUCUGGAAGGGACACACACCCUUCGGAUCGCUCCGAAG ....(((((((((((((.....))))).))))))))((..(.((.(((.........))).))....)..)).......(((((((....))))))) ( -35.10, z-score = -1.96, R) >droEre2.scaffold_4770 5395419 97 + 17746568 GUAACCUCACCCUGAGUUUAGAAUUCAGGACUGAGAGUUUCGAGUGCCAAAGUGCGAGGCUCUGGAAGGGACACACAUCCUUCGGAUCGCUCCGAAG ((..(((..((((((((.....)))))))....(((((((((...((....)).))))))))))..)))...)).....(((((((....))))))) ( -31.60, z-score = -1.17, R) >droYak2.chr3R 13597646 97 - 28832112 GUAACCUCAUCCUGAGUUUAGAAUUCAGGCCUGAGAGUUUCGAGUGCCAAAGUGCGAGGCUCUGGAAGGGACACACACUCUUCGGAUCGCUCCGAAG ((..(((((.(((((((.....)))))))..))))...(((.((.(((.........))).)).))))..)).......(((((((....))))))) ( -30.10, z-score = -0.52, R) >droSec1.super_0 8404972 97 + 21120651 GUAACCUCAGCCUGAGUUUAGAAUUCAGGCCUGAGGGCUUCGAGUGCCAAAGUGCGAGGCUCUGGAAGGGACACACACCCUUCGGAUCGCUCCGAAG ....(((((((((((((.....))))))).)))))).((((.((.(((.........))).)).))))...........(((((((....))))))) ( -40.60, z-score = -2.82, R) >consensus GUAACCUCAGCCUGAGUUUAGAAUUCAGGACUGAGAGUUUCGAGUGCCAAAGUGCGAGGCUCUGGAAGGGACACACACCCUUCGGAUCGCUCCGAAG ....(((((((((((((.....))))))).)))))).((((.((.(((.........))).)).))))...........(((((((....))))))) (-31.30 = -31.92 + 0.63)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:11:41 2011