| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,269,543 – 9,269,647 |
| Length | 104 |
| Max. P | 0.906088 |

| Location | 9,269,543 – 9,269,647 |
|---|---|
| Length | 104 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 86.03 |
| Shannon entropy | 0.27888 |
| G+C content | 0.40708 |
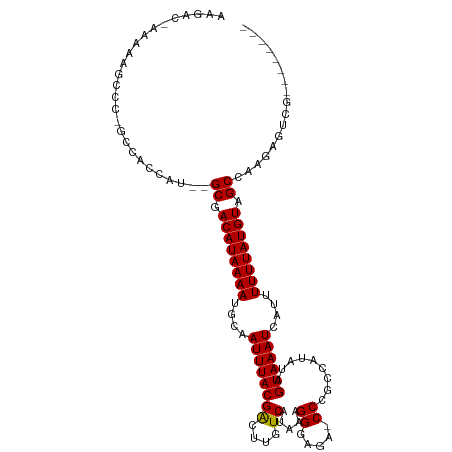
| Mean single sequence MFE | -22.40 |
| Consensus MFE | -17.32 |
| Energy contribution | -17.24 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.906088 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9269543 104 - 27905053 AAGACAAAAAAGCCC-GCCACCAU--GCGACAUAAAAUGCAAUUUACGACUUGUCAUAAAGGGAGA-CCCGCCAUAUAGUAAAUCAUUUUUUAUGUAGCCAAGAGUCG-------- ..(((.........(-((......--)))((((((((....((((((((....)).....((....-)).........))))))....))))))))........))).-------- ( -21.50, z-score = -2.56, R) >droSim1.chr3R 15354885 104 + 27517382 AAGACAAAAAAGCCC-GCCACCAU--GCGACAUAAAAUGCAAUUUACGACUUGUCAUAAAGGGAGA-CCCGCCAUAUAGUAAAUCAUUUUUUAUGUAGCCAAGAGUCG-------- ..(((.........(-((......--)))((((((((....((((((((....)).....((....-)).........))))))....))))))))........))).-------- ( -21.50, z-score = -2.56, R) >droSec1.super_0 8384800 104 + 21120651 AAGACAAAAAAGCCC-GCCACCAU--GCGACAUAAAAUGCAAUUUACGACUUGUCAUAAAGGGAGA-CCCGCCAUAUAGUAAAUCAUUUUUUAUGUAGCCAAGAGUCG-------- ..(((.........(-((......--)))((((((((....((((((((....)).....((....-)).........))))))....))))))))........))).-------- ( -21.50, z-score = -2.56, R) >droYak2.chr3R 13577083 103 - 28832112 AAGAC-AAAAAGCCC-GCCACCAU--GCGACAUAAAAUGCAAUUUACGACUUGUCAUAAAGGGAGA-CCCGCCAUAUAGUAAAUCAUUUUUUAUGUAGCCAGGAGUCG-------- ..(((-.....((.(-((......--)))((((((((....((((((((....)).....((....-)).........))))))....)))))))).)).....))).-------- ( -21.50, z-score = -1.86, R) >droEre2.scaffold_4770 5375115 104 + 17746568 AAGACAAAAAAGCCC-GCCACCAU--GCGACAUAAAAUGCAAUUUACGACUUGUCAUAAAGGGAGA-CCCGCCAUAUAGUAAAUCAUUUUUUAUGUAGCCAGGAGUCG-------- ..(((.........(-((......--)))((((((((....((((((((....)).....((....-)).........))))))....))))))))........))).-------- ( -21.50, z-score = -1.88, R) >droAna3.scaffold_13340 1828538 101 - 23697760 AAGAC-AAAAAGCCC-GCCACCAU--GCGACAUAAAAUGCAAUUUACGGCUUGUCAUAAAGGGAGA-CCCGCCAUAUAGUAAAUCAUUUUUUAUGUAGCCAG--GUCG-------- ..(((-.....((.(-((......--)))((((((((....(((((((((..(((.........))-)..))).....))))))....)))))))).))...--))).-------- ( -21.10, z-score = -1.03, R) >dp4.chr2 16716527 111 - 30794189 AAGAC-AAAAAGCCA-GCGAAGAU--GCGACAUAAAAUGCAAUUUACGACUUGUCAUAAAGGGAGA-CCCGCCAUAUAGUAAAUCAUUUUUUAUGUAGCCAAGUCGCGAGCGGAGA .....-.....((..-((((....--((.((((((((....((((((((....)).....((....-)).........))))))....)))))))).))....))))..))..... ( -28.20, z-score = -2.88, R) >droPer1.super_0 8143536 111 + 11822988 AAGAC-AAAAAGCCA-GCGAAGAU--GCGACAUAAAAUGCAAUUUACGACUUGUCAUAAAGGGAGA-CCCGCCAUAUAGUAAAUCAUUUUUUAUGUAGCCAAGUCGCGAGCGGAGA .....-.....((..-((((....--((.((((((((....((((((((....)).....((....-)).........))))))....)))))))).))....))))..))..... ( -28.20, z-score = -2.88, R) >droWil1.scaffold_181130 6342352 106 - 16660200 AAAAUGAAAACUCCAGGCCAAGAUGCGCGACAUAAAAUGCAAUUUACGACUUGUCAUAAAGGGAGA-CCCACCAUAUAGUAAAUCAUUUUUUAUGUAGCCCA-AGUCA-------- .....................(((..((.((((((((....((((((((....)).....((....-)).........))))))....)))))))).))...-.))).-------- ( -19.90, z-score = -1.24, R) >droVir3.scaffold_12855 7279233 102 + 10161210 --AAGUGUAGCUCGA-CCAAAGAU--GCGACAUAAAAUGCAAUUUACGACUUGUCAUAAAGGGAGAGCCCGCCAUAUAGUAAAUCAUUUUUUAUGUAGCCAGCUCGC--------- --..(((.(((((..-.....)).--((.((((((((....((((((((....)).....(((....)))........))))))....)))))))).))..))))))--------- ( -21.60, z-score = -0.86, R) >droMoj3.scaffold_6540 10325569 97 - 34148556 --AAGUGUAGCCCGA----AAGAU--GCGACAUAAAAUGCAAUUUACGACUUGUCAUAAAGGGAGAGCCCGCCAUAUAGUAAAUCAUUUUUUAUGUAGCGAGUUC----------- --..........(..----..).(--((.((((((((....((((((((....)).....(((....)))........))))))....)))))))).))).....----------- ( -19.90, z-score = -1.29, R) >consensus AAGAC_AAAAAGCCC_GCCACCAU__GCGACAUAAAAUGCAAUUUACGACUUGUCAUAAAGGGAGA_CCCGCCAUAUAGUAAAUCAUUUUUUAUGUAGCCAAGAGUCG________ ..........................((.((((((((....((((((((....)).....(((....)))........))))))....)))))))).))................. (-17.32 = -17.24 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:11:39 2011