
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,268,906 – 9,269,005 |
| Length | 99 |
| Max. P | 0.968772 |

| Location | 9,268,906 – 9,269,005 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 93.18 |
| Shannon entropy | 0.11601 |
| G+C content | 0.58958 |
| Mean single sequence MFE | -37.04 |
| Consensus MFE | -34.02 |
| Energy contribution | -33.70 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.837234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
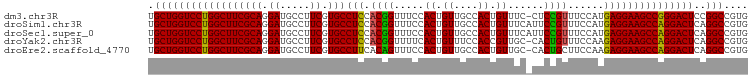
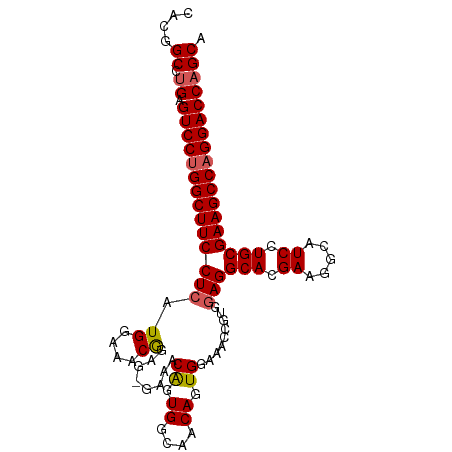
>dm3.chr3R 9268906 99 + 27905053 UGCUGGUCCUGGCUUCGCAGGAUGCCUUCGUGCCUCCACGGUUUCCACUGUUGCCACUGUUUC-CUCCGUUUCCAUGAGGAAGCCGGGACUCCGGCCGUG .(((((((((((((((((((((.((......)).)))(((((....)))))))).........-(((.........)))))))))))))..))))).... ( -36.70, z-score = -1.39, R) >droSim1.chr3R 15354250 100 - 27517382 UGCUGGUCCUGGCUUCGCAGGAUGCCUUCGUGCCUCCACGGUUUCCACUGUUGCCACUGUUUCAUUCCGUUUCCAUGAGGAAGCCAGGACUCAGGCCGUG ..((((((((((((((((((((.((......)).)))(((((....))))))))......(((((.........)))))))))))))))).)))...... ( -37.00, z-score = -1.84, R) >droSec1.super_0 8384158 100 - 21120651 UGCUGGUCCUGGCUUCGCAGGAUGCCUUCGUGCCUCCACGGUUUCCACUGUUGCCACUGUUUCAUUCCGUUUCCAUGAGGAAGCCAGGACUCAGGCCGUG ..((((((((((((((((((((.((......)).)))(((((....))))))))......(((((.........)))))))))))))))).)))...... ( -37.00, z-score = -1.84, R) >droYak2.chr3R 13576445 99 + 28832112 UGCUGGUCCUGGCUUCGCAGGAUGCCUUCGUGCCUCCACGGUUUUCACUGUUUCCACCGUUGC-CACUGUUUCCAAGAGGAAGCCAGGACUCAGGCCGUG ..((((((((((((((((((((.((......)).)))(((((.............))))))))-..((.......))..))))))))))).)))...... ( -36.82, z-score = -1.66, R) >droEre2.scaffold_4770 5374377 99 - 17746568 UGCUGGUCCUGGCUUCGCAGGAUGCCUUCGUGCCUUCACAGUUUCCACUGUUGCCACUGUUGC-CACUGCUUCCAAGAGGAAGCCAGGACUCAGGCCGUG ..((((((((((((((((((...((..(.(((.(...(((((....))))).).))).)..))-..))))(((...)))))))))))))).)))...... ( -37.70, z-score = -1.38, R) >consensus UGCUGGUCCUGGCUUCGCAGGAUGCCUUCGUGCCUCCACGGUUUCCACUGUUGCCACUGUUUC_CUCCGUUUCCAUGAGGAAGCCAGGACUCAGGCCGUG .((((((((((((((((((.((.....)).)))....(((((....)))))............................))))))))))))..))).... (-34.02 = -33.70 + -0.32)
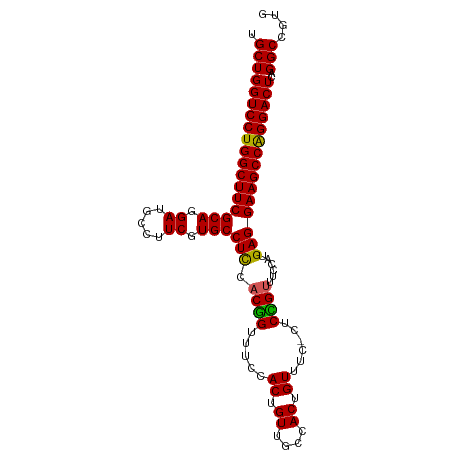
| Location | 9,268,906 – 9,269,005 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 93.18 |
| Shannon entropy | 0.11601 |
| G+C content | 0.58958 |
| Mean single sequence MFE | -41.46 |
| Consensus MFE | -36.36 |
| Energy contribution | -36.56 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.968772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
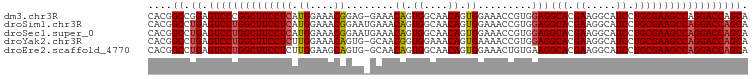
>dm3.chr3R 9268906 99 - 27905053 CACGGCCGGAGUCCCGGCUUCCUCAUGGAAACGGAG-GAAACAGUGGCAACAGUGGAAACCGUGGAGGCACGAAGGCAUCCUGCGAAGCCAGGACCAGCA (((((((((....)))))((((((.((....)))))-))).((.((....)).)).....))))..((..(...(((.((....)).))).)..)).... ( -38.50, z-score = -1.54, R) >droSim1.chr3R 15354250 100 + 27517382 CACGGCCUGAGUCCUGGCUUCCUCAUGGAAACGGAAUGAAACAGUGGCAACAGUGGAAACCGUGGAGGCACGAAGGCAUCCUGCGAAGCCAGGACCAGCA ....((.((.((((((((((((((......((((.......((.((....)).))....)))).)))(((.((.....)).)))))))))))))))))). ( -40.60, z-score = -2.62, R) >droSec1.super_0 8384158 100 + 21120651 CACGGCCUGAGUCCUGGCUUCCUCAUGGAAACGGAAUGAAACAGUGGCAACAGUGGAAACCGUGGAGGCACGAAGGCAUCCUGCGAAGCCAGGACCAGCA ....((.((.((((((((((((((......((((.......((.((....)).))....)))).)))(((.((.....)).)))))))))))))))))). ( -40.60, z-score = -2.62, R) >droYak2.chr3R 13576445 99 - 28832112 CACGGCCUGAGUCCUGGCUUCCUCUUGGAAACAGUG-GCAACGGUGGAAACAGUGAAAACCGUGGAGGCACGAAGGCAUCCUGCGAAGCCAGGACCAGCA ....((.((.(((((((((((...(((....)))..-(((((((((....).......)))))(((.((......)).))))))))))))))))))))). ( -41.91, z-score = -2.55, R) >droEre2.scaffold_4770 5374377 99 + 17746568 CACGGCCUGAGUCCUGGCUUCCUCUUGGAAGCAGUG-GCAACAGUGGCAACAGUGGAAACUGUGAAGGCACGAAGGCAUCCUGCGAAGCCAGGACCAGCA ....((.((.((((((((((((....(((.((..((-....))(((.(.(((((....)))))...).)))....)).))).).))))))))))))))). ( -45.70, z-score = -3.07, R) >consensus CACGGCCUGAGUCCUGGCUUCCUCAUGGAAACGGAG_GAAACAGUGGCAACAGUGGAAACCGUGGAGGCACGAAGGCAUCCUGCGAAGCCAGGACCAGCA ....((.((.((((((((((((((.((....))........((.((....)).)).........)))(((.((.....)).)))))))))))))))))). (-36.36 = -36.56 + 0.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:11:38 2011