
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,259,942 – 9,260,046 |
| Length | 104 |
| Max. P | 0.672142 |

| Location | 9,259,942 – 9,260,046 |
|---|---|
| Length | 104 |
| Sequences | 11 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 64.59 |
| Shannon entropy | 0.75287 |
| G+C content | 0.41438 |
| Mean single sequence MFE | -23.93 |
| Consensus MFE | -10.00 |
| Energy contribution | -10.04 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.10 |
| Mean z-score | -0.76 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.672142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9259942 104 + 27905053 AAUUAUCCACAUAGUUGUUGCGCACCUUGCGACGGCUUAGAAAACGGAGCAUUUUUCUGGAUUUUGUU---UGGUUUUAAACGUGU-------GUAUGUAUGUAUUAUAUAUUG .......(((((((((((((((.....))))))))))...(((((.(((((.............))))---).)))))....))))-------)((((((.....))))))... ( -24.42, z-score = -1.23, R) >droMoj3.scaffold_6540 10314425 79 + 34148556 CAUUAUCCGCAUAGUUGUUGCGCACCUUGCGACGACUUAGAAAACGCAGCAUUUUU-UGCAGUCGCUCGC-UUCUACACUA--------------------------------- .......((((.......))))......((((((((((((((((.......)))))-)).))))).))))-..........--------------------------------- ( -18.00, z-score = -0.73, R) >droVir3.scaffold_12855 7270417 78 - 10161210 CAUUAUCCGGAUAGUUGUUGCGCACCUUGCGCCGACUUAGAAAACGCAGCAUUUUU-UGCAGUCCGUCG--UUCUACCCAA--------------------------------- .......((((((((((..((((.....)))))))))...........(((.....-))).)))))...--..........--------------------------------- ( -18.60, z-score = -1.36, R) >droWil1.scaffold_181130 6331014 86 + 16660200 CACUAUCCGCAUAGUUGUUGCGUACCUUGCGACGGCUUAGAAAACGCAGCAUUUUG-UGGUUAGCGUUGU-UGCUGCCUUCUAUUUAG-------------------------- ............((((((((((.....))))))))))(((((...((((((...((-(.....)))....-)))))).))))).....-------------------------- ( -24.60, z-score = -1.17, R) >droSim1.chr3R 15343976 111 - 27517382 AAUUAUCCACAUAGUUGUUGCGCACCUUGCGACGGCUUAGAAAACGGAGCAUUUUUCUGGAUUUUGUU---UGGUUUUAAACGUGUGUAUUAUGUAUGUAUGUAUUAUAUAGUG .......(((((((((((((((.....))))))))))...(((((.(((((.............))))---).)))))....)))))(((((((((.........))))))))) ( -26.02, z-score = -1.08, R) >droSec1.super_0 8375005 111 - 21120651 AAUUAUCCACAUAGUUGUUGCGCACCUUGCGACGGCUUAGAAAACGGAGCAUUUUUCUGGAUUUUGUU---UGGUUUUAAACGUGUGUGUUAUGUAUGUAUGUAUUAUAUAGUG .......(((((((((((((((.....)))))))))....(((((.(((((.............))))---).))))).....)))))).....((((((.....))))))... ( -25.12, z-score = -0.57, R) >droEre2.scaffold_4770 5365116 108 - 17746568 AAUUAUCCACAUAGUUGUUGCGCACCUUGCGACGGCUUAGAAAACGGAGCAUUUUUCUGGAUUUUGUUGUUUGGUUUUAAACGUGUC-----UAU-UAUUUGUAUUAUAUAGUG ...((..(((..((((((((((.....))))))))))...(((((.(((((................))))).)))))....)))..-----)).-.................. ( -20.39, z-score = 0.07, R) >droYak2.chr3R 13567026 111 + 28832112 AAUUAUCCACAUAGUUGUUGCGCACCUUGCGACGGCUUAGAAAACGGAGCAUUUUUCUGGAUUUUGUUGUUUGGUUUUAAACGUGU---CUAUAUAUGUGUGUAUUAUAUAGUG ...((..(((((((((((((((.....)))))))))....(((((.(((((................))))).)))))........---.....))))))..)).......... ( -24.09, z-score = -0.45, R) >droAna3.scaffold_13340 1818444 109 + 23697760 CAUUAUCCGCGUAAUUGUUGCGCACCUUGCGACGGCUUAAAAAACGCAGCAUUUUCGCUGG--CUACUUUGUAUUUUUUACUUCCACCU-UUUUUGUUGUAUGAUGAUGGAA-- (((((((.(((((.....)))))....((((((((...........((((......)))).--.......(((.....)))........-...)))))))).)))))))...-- ( -24.10, z-score = -0.92, R) >dp4.chr2 16706233 109 + 30794189 CAUUAUCCGCGUAGUUGUUGCGCACCUUGCGACGGCUCAGAAAACGCAACAUUUUCGUUAUAUUUGCUUUUUCUCGGUGGCCGCUACUUGUUGUUGUCGU-CAAUGGUGG---- ......(((((.((((((((((.....)))))))))))((((((.((((..............)))).))))))..))))((((((.(((.((....)).-)))))))))---- ( -28.94, z-score = -0.47, R) >droPer1.super_0 8133025 109 - 11822988 CAUUAUCCGCGUAGUUGUUGCGCACCUUGCGACGGCUCAGAAAACGCAACAUUUUCGUUAUAUUUGCUUUUUCUCGGUGGCCGCUACUUGUUGUUGUCGU-CAAUGGUGG---- ......(((((.((((((((((.....)))))))))))((((((.((((..............)))).))))))..))))((((((.(((.((....)).-)))))))))---- ( -28.94, z-score = -0.47, R) >consensus CAUUAUCCGCAUAGUUGUUGCGCACCUUGCGACGGCUUAGAAAACGCAGCAUUUUUCUGGAUUUUGUUGU_UGGUUUUAAACGUGU____U_U_U_UGUU_GUAUUAUAUA___ ............((((((((((.....))))))))))............................................................................. (-10.00 = -10.04 + 0.03)
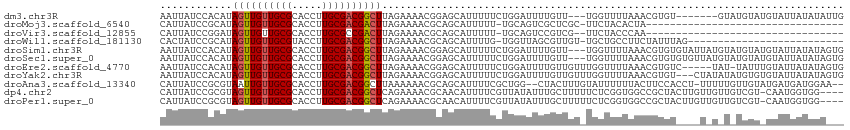

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:11:36 2011