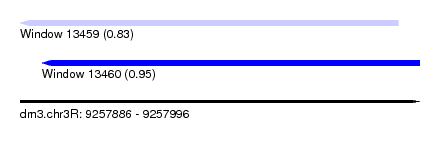
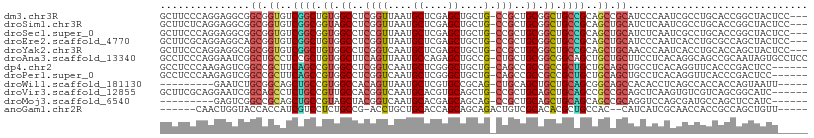
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,257,886 – 9,257,996 |
| Length | 110 |
| Max. P | 0.945020 |

| Location | 9,257,886 – 9,257,990 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 64.65 |
| Shannon entropy | 0.74794 |
| G+C content | 0.65632 |
| Mean single sequence MFE | -45.46 |
| Consensus MFE | -15.67 |
| Energy contribution | -15.36 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.830062 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
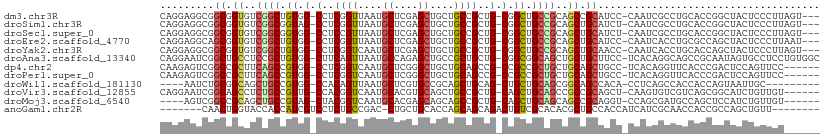
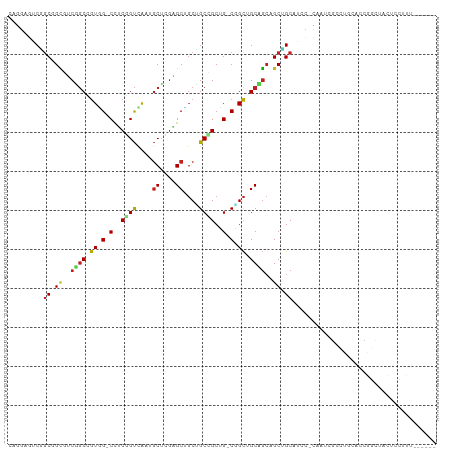
>dm3.chr3R 9257886 104 - 27905053 CAGGAGGCGGCGGUGUCGGCUGUGG-CCUCGGUUAAUGCUCGAGCUGCUGCCGCUG-CGGCUGCCGCAGCCGCAUCC-CAAUCGCCUGCACCGGCUACUCCCUUAGU--- ..((((..((.((((.(((((((((-(((((((....)).))))..(((((....)-)))).)))))))))))))))-)....(((......)))..))))......--- ( -51.20, z-score = -1.46, R) >droSim1.chr3R 15341922 104 + 27517382 CAGGAGGCGGCGGUGUCGGCGGUAG-CCUCGGUUAAUGCUCGAGCUGCUGCCGCUG-CGGCUGCCGCAGCUGCAUCU-CAAUCGCCUGCACCGGCUACUCCCUUAGU--- ..((((..(.((((((.((((((((-(.(((((....)).))))))))(((.((((-(((...))))))).)))...-....)))).)))))).)..))))......--- ( -54.60, z-score = -2.70, R) >droSec1.super_0 8372926 104 + 21120651 CAGGAGGCGGCGGUGUCGGCGGUGG-CCUCCGUUAAUGCUCGAGCUGCUGCCGCUG-CGGCUGCCGCAGCUGCAUCU-CAAUCGCCUGCACCGGCUACUCCCUUAGU--- ..((((..(.((((((.(((((..(-(.((.((....))..))))..)(((.((((-(((...))))))).)))...-....)))).)))))).)..))))......--- ( -49.60, z-score = -1.17, R) >droEre2.scaffold_4770 5363028 104 + 17746568 CAGGAGGCAGCGGUGUCGGCUGUGG-CCUCGGUUAAUGCUCGAGCUGCUGCCGCUG-CGGCUGCCGCAGCUGCAUCC-CAAUCACCUGCGCCAGCUACUCCCUUAAU--- ..(((((((((((((.(((((..((-(..........)))..))))).))))))))-)(((((.(((((.((.((..-..)))).))))).))))).))))......--- ( -47.00, z-score = -1.37, R) >droYak2.chr3R 13564889 104 - 28832112 CAGGAGGCGGCGGUGUCGGCUGUGG-CCUCGGUCAAUGCUCGAGCUGCUGCCGCUG-CGGCUGCCGCAGCUGCAACC-CAAUCACCUGCACCAGCUACUCCCUUAGU--- ((((.(((((((((.(((((..(((-(....))))..)).))))))))))))((((-(((...))))))).......-......)))).....((((......))))--- ( -45.50, z-score = -0.46, R) >droAna3.scaffold_13340 1816525 107 - 23697760 CAGGAAUCGGCUGCCUCCGCUGUGG-CUUCAGUUAAUGCCAGAGCUGCCGCUGCUG-CGGCGGCAGCUGCUGCUUCC-UCACAGGCAGCCGCAAUAGUGCCUCCUGUGGC .(((((.((((((((...(((.(((-(..........)))).))).(((((....)-))))))))))))....))))-)((((((.((.(((....))).)))))))).. ( -54.90, z-score = -2.41, R) >dp4.chr2 16704399 101 - 30794189 CAAGAGUCGGCCGCUUCAGCCGUGG-CCUCGGUCAAUGCUCGGGCUGCUGCAGCCG-CCGCCGCUGCUGCAGCUGCC-UCACAGGUUCACCCGACUCCAGUUCC------ ...(((((((((((.......))))-))..(((.(((.((.((((.((((((((.(-(....)).)))))))).)))-)...))))).))).))))).......------ ( -46.60, z-score = -2.31, R) >droPer1.super_0 8131180 101 + 11822988 CAAGAGUCGGCCGCUUCAGCCGUGG-CCUCGGUCAAUGCUCGGGCUGCUGCAGCCG-CCGCCGCUGCUGCAGCUGCC-UCACAGGUUCACCCGACUCCAGUUCC------ ...(((((((((((.......))))-))..(((.(((.((.((((.((((((((.(-(....)).)))))))).)))-)...))))).))).))))).......------ ( -46.60, z-score = -2.31, R) >droWil1.scaffold_181130 6328792 94 - 16660200 ----AAUCUGCGGCAGCUGCCGUGG-CCACAGUUAAUGCUCGUGCCGCAGCUGCAG-CUGCUGCAGCGGCAGCCACA-CCUCAGCCACCACCAGUAAUUGC--------- ----...(((((((((((((.((((-(.(((((....))).)))))))....))))-))))))))).(((.......-.....)))...............--------- ( -39.60, z-score = -1.03, R) >droVir3.scaffold_12855 7268273 101 + 10161210 CAGGAAUCGGCAGCCUCUGCCGUUG-CCACGGUCAAUGCACGUGCAGCUGCCGCUG-CAGCUGCAGCCGCCGCAGCU-CAAGUGUCGUCAGCGGCAUCUGUUGU------ ..((...((((((...))))))...-))..(((...((((..((((((....))))-))..))))...)))(((((.-...((((((....))))))..)))))------ ( -43.20, z-score = -0.44, R) >droMoj3.scaffold_6540 10312336 97 - 34148556 ----AGUCGGCCGCAGCUGCCGUAG-CUACGGUCAAUGCACGAGCAGCAGCCGCUG-CAGCUGCAGCAGCCGCAGGU-CCAGCGAUGCCAGCUCCAUCUGUUGU------ ----....((((((.(((((.((((-((.((((....)).)).(((((....))))-))))))).))))).)).)))-)((((((((.......)))).)))).------ ( -41.90, z-score = -0.66, R) >anoGam1.chr2R 52445529 94 - 62725911 -------CAACUGGUACCACCAUCGUCCUCUGCCGAC-CUGCUGCACCAGCAGCAGACUGUCGCACACGCUGCCACCAUCAUCGCAACCACCGCCAGCUGUU-------- -------((.(((((........(((....(((.(((-(((((((....)))))))...)))))).))).(((..........)))......))))).))..-------- ( -24.80, z-score = -1.09, R) >consensus CAGGAGUCGGCGGCGUCGGCCGUGG_CCUCGGUCAAUGCUCGAGCUGCUGCCGCUG_CGGCUGCAGCAGCUGCAUCC_CAAUCGCCUGCACCGGCUACUCCUUU______ .........((.((..((((.((.(.(..((((....((....))....))))..).).)).))))..)).))..................................... (-15.67 = -15.36 + -0.31)
| Location | 9,257,892 – 9,257,996 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 65.35 |
| Shannon entropy | 0.73904 |
| G+C content | 0.67176 |
| Mean single sequence MFE | -45.77 |
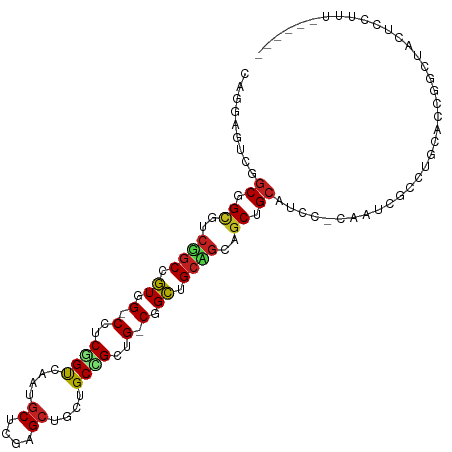
| Consensus MFE | -16.56 |
| Energy contribution | -16.42 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.945020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9257892 104 - 27905053 GCUUCCCAGGAGGCGGCGGUGUCGGCUGUGGCCUCGGUUAAUGCUCGAGCUGCUG-CCGCUGCGGCUGCCGCAGCCGCAUCCCAAUCGCCUGCACCGGCUACUCC--- (((((....)))))((.((((.((((((((((((((((....)).))))..((((-(....))))).))))))))))))))))....(((......)))......--- ( -52.70, z-score = -2.02, R) >droSim1.chr3R 15341928 104 + 27517382 GCUUCUCAGGAGGCGGCGGUGUCGGCGGUAGCCUCGGUUAAUGCUCGAGCUGCUG-CCGCUGCGGCUGCCGCAGCUGCAUCUCAAUCGCCUGCACCGGCUACUCC--- (((((....)))))(.((((((.(((((((((.(((((....)).))))))))((-(.(((((((...))))))).))).......)))).)))))).)......--- ( -54.10, z-score = -2.76, R) >droSec1.super_0 8372932 104 + 21120651 GCUUCCCAGGAGGCGGCGGUGUCGGCGGUGGCCUCCGUUAAUGCUCGAGCUGCUG-CCGCUGCGGCUGCCGCAGCUGCAUCUCAAUCGCCUGCACCGGCUACUCC--- (((((....)))))(.((((((.(((((..((.((.((....))..))))..)((-(.(((((((...))))))).))).......)))).)))))).)......--- ( -50.20, z-score = -1.37, R) >droEre2.scaffold_4770 5363034 104 + 17746568 GCUUCGCAGGAGGCAGCGGUGUCGGCUGUGGCCUCGGUUAAUGCUCGAGCUGCUG-CCGCUGCGGCUGCCGCAGCUGCAUCCCAAUCACCUGCGCCAGCUACUCC--- (((.((((((.(((((((((.(((((..((((....))))..)).))))))))))-))(((((((...))))))).............))))))..)))......--- ( -52.70, z-score = -2.14, R) >droYak2.chr3R 13564895 104 - 28832112 GCUUCCCAGGAGGCGGCGGUGUCGGCUGUGGCCUCGGUCAAUGCUCGAGCUGCUG-CCGCUGCGGCUGCCGCAGCUGCAACCCAAUCACCUGCACCAGCUACUCC--- (((...((((.(((((((((.(((((..((((....))))..)).))))))))))-))(((((((...))))))).............))))....)))......--- ( -47.90, z-score = -1.20, R) >droAna3.scaffold_13340 1816531 107 - 23697760 GCCUCCCAGGAAUCGGCUGCCUCCGCUGUGGCUUCAGUUAAUGCCAGAGCUGCCG-CUGCUGCGGCGGCAGCUGCUGCUUCCUCACAGGCAGCCGCAAUAGUGCCUCC ((((...(((((.((((((((...(((.((((..........)))).))).((((-(....)))))))))))))....)))))...))))((.(((....))).)).. ( -50.20, z-score = -1.90, R) >dp4.chr2 16704405 101 - 30794189 GCCUCCCAAGAGUCGGCCGCUUCAGCCGUGGCCUCGGUCAAUGCUCGGGCUGCUG-CAGCCGCCGCCGCUGCUGCAGCUGCCUCACAGGUUCACCCGACUCC------ .........(((((((((((.......))))))..(((.(((.((.((((.((((-((((.((....)).)))))))).))))...))))).))).))))).------ ( -46.60, z-score = -2.27, R) >droPer1.super_0 8131186 101 + 11822988 GCCUCCCAAGAGUCGGCCGCUUCAGCCGUGGCCUCGGUCAAUGCUCGGGCUGCUG-CAGCCGCCGCCGCUGCUGCAGCUGCCUCACAGGUUCACCCGACUCC------ .........(((((((((((.......))))))..(((.(((.((.((((.((((-((((.((....)).)))))))).))))...))))).))).))))).------ ( -46.60, z-score = -2.27, R) >droWil1.scaffold_181130 6328794 93 - 16660200 ---------GAAUCUGCGGCAGCUGCCGUGGCCACAGUUAAUGCUCGUGCCGCAG-CUGCAGCUGCUGCAGCGGCAGCCACACCUCAGCCACCACCAGUAAUU----- ---------....(((((((((((((.(((((.(((((....))).)))))))..-..))))))))))))).(((............))).............----- ( -39.60, z-score = -1.60, R) >droVir3.scaffold_12855 7268279 101 + 10161210 GCUUCGCAGGAAUCGGCAGCCUCUGCCGUUGCCACGGUCAAUGCACGUGCAGCUG-CCGCUGCAGCUGCAGCCGCCGCAGCUCAAGUGUCGUCAGCGGCAUC------ (((..((.((...((((((...))))))...))..(((...((((..((((((..-..))))))..))))...))))))))....((((((....)))))).------ ( -42.80, z-score = -0.26, R) >droMoj3.scaffold_6540 10312342 92 - 34148556 ---------GAGUCGGCCGCAGCUGCCGUAGCUACGGUCAAUGCACGAGCAGCAG-CCGCUGCAGCUGCAGCAGCCGCAGGUCCAGCGAUGCCAGCUCCAUC------ ---------((((.(((.((.(((((.((((((.((((....)).)).(((((..-..))))))))))).))))).))..(((....)))))).))))....------ ( -41.10, z-score = -1.36, R) >anoGam1.chr2R 52445529 94 - 62725911 ------CAACUGGUACCACCAUCGUCCUCUGCCG-ACCUGCUGCACCAGCAGCAGACUGUCGCACACGCUGCCAC--CAUCAUCGCAACCACCGCCAGCUGUU----- ------((.(((((........(((....(((.(-(((((((((....)))))))...)))))).))).(((...--.......)))......))))).))..----- ( -24.80, z-score = -1.09, R) >consensus GCUUCCCAGGAGGCGGCGGCGUCGGCCGUGGCCUCGGUUAAUGCUCGAGCUGCUG_CCGCUGCGGCUGCCGCAGCUGCAUCCCAACCGCCUGCACCGGCUACU_____ ...............((.((..((((.((.((..(((.....((....))......)))..)).)).))))..)).)).............................. (-16.56 = -16.42 + -0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:11:35 2011