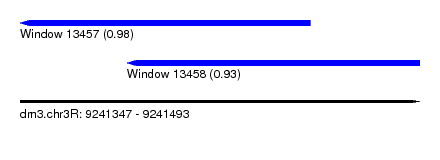
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,241,347 – 9,241,493 |
| Length | 146 |
| Max. P | 0.981240 |

| Location | 9,241,347 – 9,241,453 |
|---|---|
| Length | 106 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 66.26 |
| Shannon entropy | 0.64055 |
| G+C content | 0.44171 |
| Mean single sequence MFE | -23.34 |
| Consensus MFE | -7.49 |
| Energy contribution | -8.00 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.981240 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9241347 106 - 27905053 AAGUGUCUGUUUACCUUAAAUUCAGCCGCAUCAUCAUGCAGUACAAUGGA----------CUCUCCAUUGACAAAA---ACAGGCACUGAUAUGCUGAUAAAAAGACACAAAAUUCACC ..((((((.((((.(......((((((((((....)))).((.((((((.----------....))))))))....---...)))..)))......).)))).)))))).......... ( -28.20, z-score = -3.79, R) >droSim1.chr3R 15325172 106 + 27517382 AACUGUCUGUUUACCUUAAAUUCCGCCGCAUCACCAUGCGGUACAAUGGA----------CUCUCCAUUGACAAAA---ACAGCCAAUGAUAUGCUGAUAAAAAUACACAAAAUUCACC ...(((.(((((............(((((((....))))))).((((((.----------....))))))......---.((((.........))))....))))).)))......... ( -23.70, z-score = -3.66, R) >droSec1.super_0 8356425 106 + 21120651 AACUGUCUGUUUACCUUAAAUUCCGCCGCAUCACCAUGCGGUACAAUGGA----------CUCUCCAUUGACAAAA---ACAGCCACUGAUAUGCUGAUAAAAAUACACAAAAUUCACG ...(((.(((((............(((((((....))))))).((((((.----------....))))))......---.((((.........))))....))))).)))......... ( -23.70, z-score = -3.79, R) >droEre2.scaffold_4770 5346636 105 + 17746568 AACUGUCUGUUUACCUUAAAUUCCGACGCAUCACCAUGCGGUACAAUGGA----------CUCUCCAUUCACAAAA---ACAGCCGCUGAUAUGCUGAUACAAACGC-CGAAAUUCACC ....(.((((((..............(((((....)))))((..(((((.----------....))))).))..))---)))).)((......))............-........... ( -16.30, z-score = -0.31, R) >droYak2.chr3R 13548400 105 - 28832112 AACUGUCUGUUUACCUUAAAUUCCGCCGCAUCACCAUGCGGUACAAUGGA----------CUCUCCAUUGACAAAA---ACAGGCGCUGAUAUGCUGAUACAAAUGC-CGAAAUCGCCC ....((((((((............(((((((....))))))).((((((.----------....))))))....))---))))))((.(((..((..........))-....))))).. ( -29.00, z-score = -3.43, R) >droAna3.scaffold_13340 1800372 90 - 23697760 AACUCCACAUAUACCUUAAAUUC----------CUGAGCAGUACAAUGGA--------CUCUCUCCAUUGACAAAA---ACGGUCGCAGCUAUAAAGGCAACAAAAUUCGC-------- ......................(----------((.(((.((.(((((((--------.....)))))))))....---..(....).)))....))).............-------- ( -13.20, z-score = -0.64, R) >dp4.chr2 16688449 104 - 30794189 -----------AGCUUUCGACACUGCC--UGCUUUCGGCAGUACAAUGGACUCUCUCUCUCCCUCCAUUGACAAAACUGCCGUCCUCGGCUUUACUCGUA--AAUACCCGAAAUUCGAG -----------.....((((....(((--......(((((((.(((((((.............))))))).....))))))).....))).....(((..--......)))...)))). ( -25.52, z-score = -3.04, R) >droPer1.super_0 8115123 102 + 11822988 -----------AGCUUUCGACACUGCC--UGCUUUCGGCAGUACAAUGGA--CUCUCUCUCCCUCCAUUGACAAAACGGCCGUCCGCGGCUUUACUCGUA--AAUACCCGAAAUUCGAG -----------...(((((..((((((--.......)))))).(((((((--...........))))))).......(((((....))))).........--......)))))...... ( -27.10, z-score = -2.30, R) >consensus AACUGUCUGUUUACCUUAAAUUCCGCCGCAUCACCAUGCAGUACAAUGGA__________CUCUCCAUUGACAAAA___ACAGCCGCUGAUAUGCUGAUAAAAAUACACGAAAUUCACC ......................((((...........))))..(((((((.............)))))))................................................. ( -7.49 = -8.00 + 0.50)
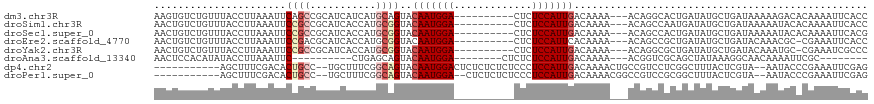
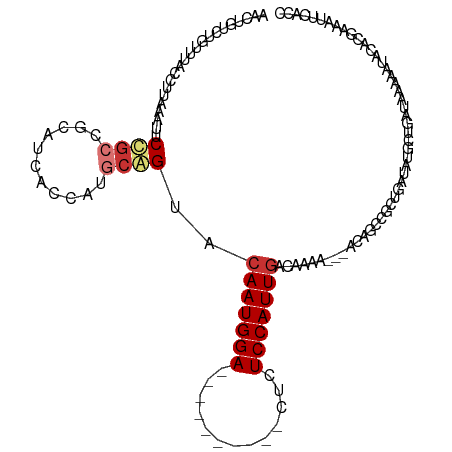
| Location | 9,241,386 – 9,241,493 |
|---|---|
| Length | 107 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.89 |
| Shannon entropy | 0.45924 |
| G+C content | 0.37802 |
| Mean single sequence MFE | -23.57 |
| Consensus MFE | -9.13 |
| Energy contribution | -9.02 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.933383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9241386 107 - 27905053 UUGUGGUAUUUUAAUUGCCAACCCAGUACAUUUAAUUUAUAAGUGUCUGUUUACCUUAAAUUCAGCCGCAUCAUCAUGCAGUACAAUGGA----------CUCUCCAUUGACAAAAA--- ...(((((.......)))))...(((.(((((((.....))))))))))..................((((....)))).((.((((((.----------....)))))))).....--- ( -23.30, z-score = -2.45, R) >droSim1.chr3R 15325211 107 + 27517382 UUGUGGUAUUUUAAUUGCCAACCCAGUACAUUUAAUUUAUAACUGUCUGUUUACCUUAAAUUCCGCCGCAUCACCAUGCGGUACAAUGGA----------CUCUCCAUUGACAAAAA--- ...(((((.......)))))...(((.(((.(((.....))).))))))...............(((((((....))))))).((((((.----------....)))))).......--- ( -25.20, z-score = -3.09, R) >droSec1.super_0 8356464 107 + 21120651 UUGUGGUAUUUUAAUUGUCAACCCAGUACAUUUAAUUUAUAACUGUCUGUUUACCUUAAAUUCCGCCGCAUCACCAUGCGGUACAAUGGA----------CUCUCCAUUGACAAAAA--- ((((((...(((((..((.(((.((((..............))))...))).)).)))))..))(((((((....))))))).((((((.----------....))))))))))...--- ( -23.84, z-score = -2.67, R) >droEre2.scaffold_4770 5346674 107 + 17746568 UUGUGGUAUUUUAAUUGCCACUCCAGUACAUUUAAUUUAUAACUGUCUGUUUACCUUAAAUUCCGACGCAUCACCAUGCGGUACAAUGGA----------CUCUCCAUUCACAAAAA--- ..((((((.......))))))....((((((((((.....(((.....)))....)))))).....(((((....)))))))))(((((.----------....)))))........--- ( -22.20, z-score = -2.63, R) >droYak2.chr3R 13548438 107 - 28832112 UUGUGGUAUUUUAAUUGCCACCCCAGUACAUUUAAUUUAUAACUGUCUGUUUACCUUAAAUUCCGCCGCAUCACCAUGCGGUACAAUGGA----------CUCUCCAUUGACAAAAA--- ..((((((.......))))))..(((.(((.(((.....))).))))))...............(((((((....))))))).((((((.----------....)))))).......--- ( -27.70, z-score = -4.01, R) >droAna3.scaffold_13340 1800403 99 - 23697760 UUAGGAUAAUUUAAAAGCCAGCGGAGAACACUUAAUUUAUAACUCCACAUAUACCUUAAAUUC----------CUGAGCAGUACAAUGGA--------CUCUCUCCAUUGACAAAAA--- ((((((..((((((..(....)((((................)))).........))))))))----------))))...((.(((((((--------.....))))))))).....--- ( -19.29, z-score = -2.70, R) >dp4.chr2 16688486 107 - 30794189 UUGCGGCAUUUUAAUUGUCGUUCGAGUACAUUUAAUUUAUAGCUUUCGACACUGCCUGCUUUC-------------GGCAGUACAAUGGACUCUCUCUCUCCCUCCAUUGACAAAACUGC ..((((..........((((...((((..............)))).))))((((((.......-------------)))))).(((((((.............)))))))......)))) ( -24.36, z-score = -2.16, R) >droPer1.super_0 8115160 105 + 11822988 UUGCGGUAUUUUAAUUGUCGUUCGAAUACAUUUAAUUUAUAGCUUUCGACACUGCCUGCUUUC-------------GGCAGUACAAUGGAC--UCUCUCUCCCUCCAUUGACAAAACGGC ..((.(((..((((.(((.........))).))))..))).))..(((..((((((.......-------------)))))).(((((((.--..........)))))))......))). ( -22.70, z-score = -1.71, R) >consensus UUGUGGUAUUUUAAUUGCCAACCCAGUACAUUUAAUUUAUAACUGUCUGUUUACCUUAAAUUCCGCCGCAUCACCAUGCAGUACAAUGGA__________CUCUCCAUUGACAAAAA___ ...(((((.......)))))............................................................((.(((((((.............)))))))))........ ( -9.13 = -9.02 + -0.11)

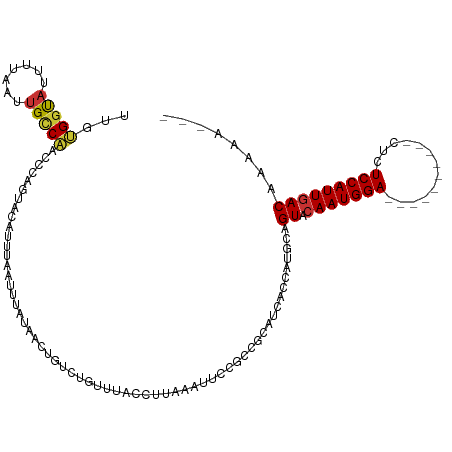
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:11:33 2011