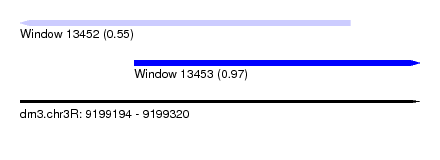
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,199,194 – 9,199,320 |
| Length | 126 |
| Max. P | 0.971931 |

| Location | 9,199,194 – 9,199,298 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 71.02 |
| Shannon entropy | 0.57142 |
| G+C content | 0.36369 |
| Mean single sequence MFE | -18.43 |
| Consensus MFE | -9.61 |
| Energy contribution | -10.31 |
| Covariance contribution | 0.71 |
| Combinations/Pair | 1.73 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.545457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
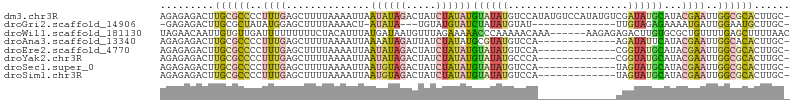
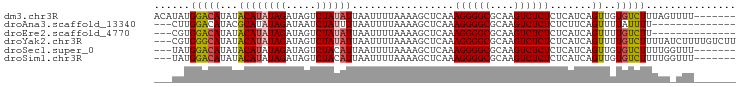
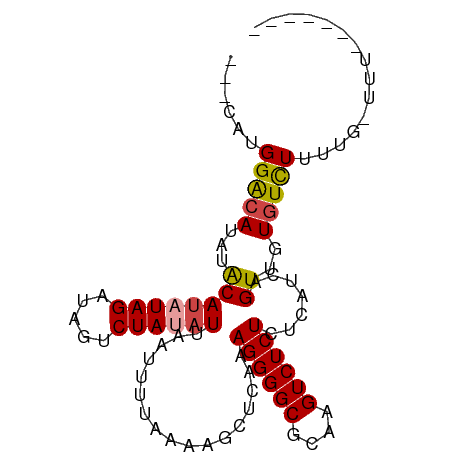
>dm3.chr3R 9199194 104 - 27905053 AGAGAGACUUGCGCCCCUUUGAGCUUUUAAAAUUAAUAUAGACUAUCUAUAUGUAUAUGUCCAUAUGUCCAUAUGUCGAUAUGCAUACGAAUUGGCGCACUUGC- .........((((((..((((..............((((((.....))))))((((((...(((((....)))))...))))))...))))..)))))).....- ( -24.70, z-score = -1.98, R) >droGri2.scaffold_14906 3719426 85 - 14172833 -GAGAGACUUGCGCUAUAUGGAGCUUUUAAAACU-AUAUA---UGUAUGUAUCUAUAUGUAU--------------UUGUAGAGAAAAUGAUUGGAAUGCUUGC- -..(((((.((((.(((((((...........))-)))))---)))).)).((((((.....--------------.))))))................)))..- ( -11.70, z-score = 0.73, R) >droWil1.scaffold_181130 6276613 99 - 16660200 UAGAACAAUUGUGUUGAUUUUUUUUUCUACAUUUAUGAUAAUGUUUAGAAAAACCCAAAAACAAA------AAGAGAGACUUGUGCGCUGUUUUGAGCUUUUAAC ...........((((......((((((((((((......)))))..)))))))......))))((------(((.(((((.........)))))...)))))... ( -13.80, z-score = 0.27, R) >droAna3.scaffold_13340 1760926 91 - 23697760 AGAGAGACUUGCGCCCCUUUGAGCUUUUAAAAUUAAAAUAGAUUAUCUAUAUGCGUAUGUCCA-------------AGAUAUUCAUACGAAUUGGCACACUUGC- .(((.(((.(((((...(((((..........)))))((((.....))))..))))).)))..-------------.(..((((....))))...)...)))..- ( -12.50, z-score = 0.57, R) >droEre2.scaffold_4770 5301042 91 + 17746568 AGAGAGACUUGCGCCCCUUUGAGCUUUUAAAAUUAAUAUAGACUAUCUAUAUGUAUAUGUCCA-------------CGGUAUGCAUACGAAUUGGCGCACUUGC- .........((((((..((((..............((((((.....))))))((((((.....-------------..))))))...))))..)))))).....- ( -20.90, z-score = -1.48, R) >droYak2.chr3R 13506863 91 - 28832112 AGAGAGACUUGCGCCCCUUUGAGCUUUUAAAAUUAAUAUAGACUAUCUAUAUGUAUAUGCCCA-------------CGGUAUGCAUACGAAUUGGCGCACUUGC- .........((((((..((((..............((((((.....))))))...((((((..-------------.))))))....))))..)))))).....- ( -21.00, z-score = -1.66, R) >droSec1.super_0 8308926 91 + 21120651 AGAGAGACUUGCGCCCCUUUGAGCUUUUAAAAUUAAUGUAGACUAUCUAUAUGUAUAUGUCCA-------------UAGUAUGCAUACGAAUUGGCGCACUUGC- .........((((((..((((..............((((((.....))))))((((((.....-------------..))))))...))))..)))))).....- ( -21.40, z-score = -1.69, R) >droSim1.chr3R 15283743 91 + 27517382 AGAGAGACUUGCGCCCCUUUGAGCUUUUAAAAUUAAUGUAGACUAUCUAUAUGUAUAUGUCCA-------------UAGUAUGCAUACGAAUUGGCGCACUUGC- .........((((((..((((..............((((((.....))))))((((((.....-------------..))))))...))))..)))))).....- ( -21.40, z-score = -1.69, R) >consensus AGAGAGACUUGCGCCCCUUUGAGCUUUUAAAAUUAAUAUAGACUAUCUAUAUGUAUAUGUCCA_____________CGGUAUGCAUACGAAUUGGCGCACUUGC_ .........((((((..((((..............((((((.....))))))((((((....................))))))...))))..))))))...... ( -9.61 = -10.31 + 0.71)
| Location | 9,199,230 – 9,199,320 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 85.46 |
| Shannon entropy | 0.25562 |
| G+C content | 0.33788 |
| Mean single sequence MFE | -17.33 |
| Consensus MFE | -15.16 |
| Energy contribution | -15.42 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.971931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
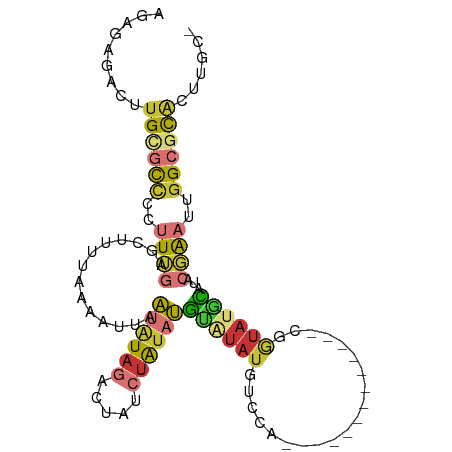
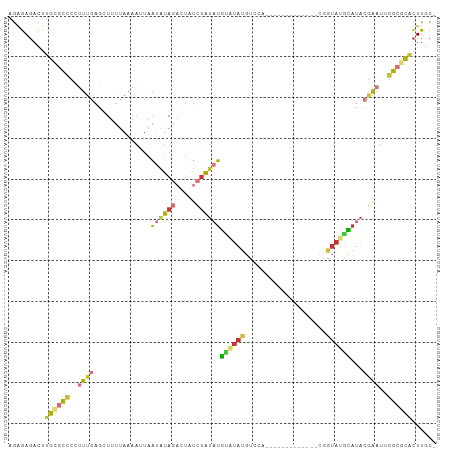
>dm3.chr3R 9199230 90 + 27905053 ACAUAUGGACAUAUACAUAUAGAUAGUCUAUAUUAAUUUUAAAAGCUCAAAGGGGCGCAAGUCUCUCUCAUCAGUUGUGUCUUUAGUUUU------- ...((.(((((((.((((((((.....)))))).................((((((....)))))).......))))))))).)).....------- ( -20.50, z-score = -3.04, R) >droAna3.scaffold_13340 1760952 80 + 23697760 ---CUUGGACAUACGCAUAUAGAUAAUCUAUUUUAAUUUUAAAAGCUCAAAGGGGCGCAAGUCUCUCUCUUCAGUUUUAUUUU-------------- ---...............((((.....))))..........((((((.((((((((....))))))...)).)))))).....-------------- ( -12.60, z-score = -1.16, R) >droEre2.scaffold_4770 5301068 80 - 17746568 ---CGUGGACAUAUACAUAUAGAUAGUCUAUAUUAAUUUUAAAAGCUCAAAGGGGCGCAAGUCUCUCUCAUCAGUUUUGUCUU-------------- ---...((((......((((((.....))))))........((((((...((((((....))))))......)))))))))).-------------- ( -18.40, z-score = -3.20, R) >droYak2.chr3R 13506889 94 + 28832112 ---CGUGGGCAUAUACAUAUAGAUAGUCUAUAUUAAUUUUAAAAGCUCAAAGGGGCGCAAGUCUCUCUCAUCAGUUUUGUCUUUUAUCUUUUGUCUU ---...(((((.....((((((.....))))))........((((((...((((((....))))))......)))))).............))))). ( -18.50, z-score = -2.29, R) >droSec1.super_0 8308952 87 - 21120651 ---UAUGGACAUAUACAUAUAGAUAGUCUACAUUAAUUUUAAAAGCUCAAAGGGGCGCAAGUCUCUCUCAUCAGUUGUGUCUUUUGGUUU------- ---...(((((((.((...(((.....)))....................((((((....)))))).......)))))))))........------- ( -17.00, z-score = -1.46, R) >droSim1.chr3R 15283769 87 - 27517382 ---UAUGGACAUAUACAUAUAGAUAGUCUACAUUAAUUUUAAAAGCUCAAAGGGGCGCAAGUCUCUCUCAUCAGUUGUGUCUUUUGGUUU------- ---...(((((((.((...(((.....)))....................((((((....)))))).......)))))))))........------- ( -17.00, z-score = -1.46, R) >consensus ___CAUGGACAUAUACAUAUAGAUAGUCUAUAUUAAUUUUAAAAGCUCAAAGGGGCGCAAGUCUCUCUCAUCAGUUGUGUCUUUUG_UUU_______ ......(((((...((((((((.....)))))).................((((((....)))))).......))..)))))............... (-15.16 = -15.42 + 0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:11:29 2011