
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,114,580 – 9,114,673 |
| Length | 93 |
| Max. P | 0.585227 |

| Location | 9,114,580 – 9,114,673 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 77.21 |
| Shannon entropy | 0.43153 |
| G+C content | 0.58500 |
| Mean single sequence MFE | -34.64 |
| Consensus MFE | -17.32 |
| Energy contribution | -17.58 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.585227 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9114580 93 + 27905053 -------------ACUGGGCGGCUACUUGAAAUUCGGGAGCCGGACAUCGCGUAGCC-------GCAGCGUAGCACGAACGUGCGCCAAAACAAGCAAAUCGUGUCGAAGCCA -------------...((((((((((.(((..(((((...)))))..))).))))))-------))..((..((((((...(((..........)))..))))))))...)). ( -35.30, z-score = -1.49, R) >droWil1.scaffold_181130 6197328 98 + 16660200 AGUUAGCGUCGACGGGGGGCAACUACUUGAAAUUCGUCAGCCGGACAUAGCAAUGUC---------------GCACGAACGUGCACCAAAACAAGCAAAUCGUGCCAGACCCA .....(.(((((((((...(((....)))...)))))).....(((((....)))))---------------((((((...(((..........)))..))))))..))).). ( -26.90, z-score = -0.35, R) >droSim1.chr3R 15206244 93 - 27517382 -------------ACUGGGCGGCUACUUGAAAUUCGGGAGCCGGACAUCGCGUAGCC-------GCAGCGUAGCACGAACGUGCGCCAAAACAAGCAAAUCGUGCCGAUGCCA -------------.....((((((((.(((..(((((...)))))..))).))))))-------)).((((.((((((...(((..........)))..))))))..)))).. ( -38.70, z-score = -2.16, R) >droSec1.super_0 8230657 93 - 21120651 -------------ACUGGGCGGCUACUUGAAAUUCGGCAGCCGGACAUCGCGUAGCC-------GCAGCGUAGCACGAACGUGCGCCAAAACAAGCAAAUCGUGCCGAUGCCA -------------.....((((((((.(((..(((((...)))))..))).))))))-------)).((((.((((((...(((..........)))..))))))..)))).. ( -38.70, z-score = -2.10, R) >droYak2.chr3R 13419379 93 + 28832112 -------------ACUGGGCGGCUACUUGAAAUUCGGGAGCCGGACAUCGCGUAGCC-------GCAGCGUGGCACGAACGUGCGCCAAAACAAGCAAGUCGUGCCGCUGCCU -------------.....((((((((.(((..(((((...)))))..))).))))))-------)).(((((((((((...(((..........)))..))))))))).)).. ( -45.20, z-score = -2.99, R) >droEre2.scaffold_4770 5212451 93 - 17746568 -------------GCUGGGCGGCUACUUGAAAUUCGGGAGCCGGACAUCGCGUAGCC-------GCAGCGUGGCACGAACGUGCGCCAAAACAAGCGAAUCGUGCCGCUGCCA -------------(((..((((((((.(((..(((((...)))))..))).))))))-------)))))(((((((((...(((..........)))..)))))))))..... ( -44.10, z-score = -2.31, R) >droVir3.scaffold_12855 7114998 71 - 10161210 -----------CUGUUGG--GGCUACUUGAAAUUCGACAGCCGGACGUCG-----------------------CACGAACGUGCGCCAAAACAAGCAAUUCGUGCCG------ -----------.((((..--(((.....((..((((.....))))..))(-----------------------(((....)))))))..)))).(((.....)))..------ ( -20.40, z-score = -0.21, R) >dp4.chr2 20804434 87 - 30794189 -------------UCUGGGCGGCUACUUGAAAUUCGACAGCCGGACAUCGGAGCGUC----------UCACGGCACGAACGUGCGCCAAAACAAGCAAAUCGUGCCACAG--- -------------(((((.(((((..(((.....))).)))))....))))).....----------....(((((((...(((..........)))..)))))))....--- ( -27.90, z-score = -1.26, R) >droPer1.super_0 10493197 86 + 11822988 -------------UCUGG-CGGCUACUUGAAAUUCGACAGCCGGACAUCGGAGCGUC----------UCACGGCACGAACGUGCGCCAAAACAAGCAAAUCGUGCCACAG--- -------------.(((.-(((((..(((.....))).)))))(((........)))----------....(((((((...(((..........)))..))))))).)))--- ( -27.40, z-score = -1.34, R) >droAna3.scaffold_13340 1669621 100 + 23697760 -------------GAUGGGCAGCUACUUGAAAUUCGGGGGCCGGACAUCGCACUCUCCGGCAAAGCAUCGUGGCACGAACGUGCGCCAAAACAAGCAAAUCGUGCCGCAGCCA -------------....(((.(((.((((.....)))).((((((..........))))))..)))...(((((((((...(((..........)))..))))))))).))). ( -41.80, z-score = -2.84, R) >consensus _____________ACUGGGCGGCUACUUGAAAUUCGGCAGCCGGACAUCGCGUAGCC_______GCAGCGUGGCACGAACGUGCGCCAAAACAAGCAAAUCGUGCCGCAGCCA ...............((..(((((...((.....))..)))))..)).........................((((((...(((..........)))..))))))........ (-17.32 = -17.58 + 0.26)

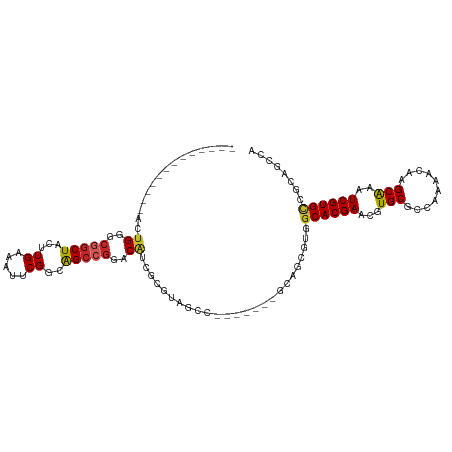
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:11:25 2011