
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,107,906 – 9,107,998 |
| Length | 92 |
| Max. P | 0.893095 |

| Location | 9,107,906 – 9,107,998 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 96.20 |
| Shannon entropy | 0.06740 |
| G+C content | 0.45217 |
| Mean single sequence MFE | -21.52 |
| Consensus MFE | -21.42 |
| Energy contribution | -21.02 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.08 |
| Mean z-score | -0.96 |
| Structure conservation index | 1.00 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.712900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9107906 92 + 27905053 UAUGAAGAUUCAUAGUGCCAUAGUGAAAUAGCAAUGCGUCCUGCAUUUAAAUGAGGGAAAGCACUGCCACAGAGGAGCACCCACAGAUAGAU (((((....)))))(((((...(((.....(((.(((.((((.(((....)))))))...))).))))))....).))))............ ( -20.30, z-score = -1.15, R) >droEre2.scaffold_4770 5205282 92 - 17746568 UAUGGAGAUUCAUAGUGCCAUAGUGAAAUAGCAAUGCGUCCUGCAUUUAAAUGAGGGAAAGCAGUGCCACAGAGGAGCACCCCCAGAUUGAU (((((....)))))(((((...(((.....(((.(((.((((.(((....)))))))...))).))))))....).))))............ ( -19.40, z-score = -0.14, R) >droYak2.chr3R 13412281 92 + 28832112 UAUGGAGAUUCAUAGUGCCAUAGUGAAAUAGCAAUGCGUCCUGCAUUUAAAUGAGGGAAAGCACUGCCACAGAGGAGCACCCUCCGAUUGAU ..(((((.......(((((...(((.....(((.(((.((((.(((....)))))))...))).))))))....).)))).)))))...... ( -21.90, z-score = -0.57, R) >droSec1.super_0 8223976 92 - 21120651 UAUGGAGAUCCAUAGUGCCAUAGUGAUAUAGCAAUGCGUCCUGCAUUUAAAUGAGGGAAAGCACUGCCACAGAGGAGCACCCACAGAUAGAU (((((....)))))(((((...(((.....(((.(((.((((.(((....)))))))...))).))))))....).))))............ ( -23.00, z-score = -1.38, R) >droSim1.chr3R 15199486 92 - 27517382 UAUGGAGAUCCAUAGUGCCAUAGUGAAAUAGCAAUGCGUCCUGCAUUUAAAUGAGGGAAAGCACUGCCACAGAGGAGCACCCACAGAUAGAU (((((....)))))(((((...(((.....(((.(((.((((.(((....)))))))...))).))))))....).))))............ ( -23.00, z-score = -1.57, R) >consensus UAUGGAGAUUCAUAGUGCCAUAGUGAAAUAGCAAUGCGUCCUGCAUUUAAAUGAGGGAAAGCACUGCCACAGAGGAGCACCCACAGAUAGAU (((((....)))))(((((...(((.....(((.(((.((((.(((....)))))))...))).))))))....).))))............ (-21.42 = -21.02 + -0.40)

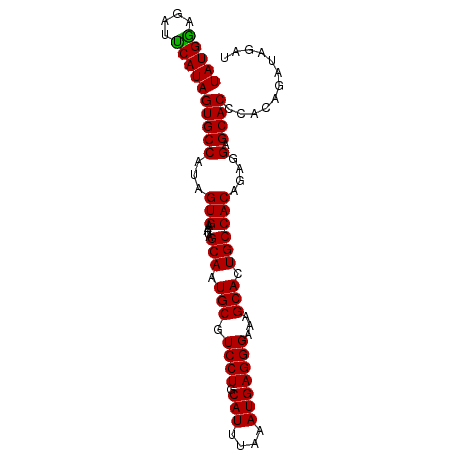

| Location | 9,107,906 – 9,107,998 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 96.20 |
| Shannon entropy | 0.06740 |
| G+C content | 0.45217 |
| Mean single sequence MFE | -26.62 |
| Consensus MFE | -24.24 |
| Energy contribution | -24.68 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.893095 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9107906 92 - 27905053 AUCUAUCUGUGGGUGCUCCUCUGUGGCAGUGCUUUCCCUCAUUUAAAUGCAGGACGCAUUGCUAUUUCACUAUGGCACUAUGAAUCUUCAUA ...........((((((.....((((((((((..(((..(((....)))..))).))))))))))........))))))((((....)))). ( -25.22, z-score = -2.03, R) >droEre2.scaffold_4770 5205282 92 + 17746568 AUCAAUCUGGGGGUGCUCCUCUGUGGCACUGCUUUCCCUCAUUUAAAUGCAGGACGCAUUGCUAUUUCACUAUGGCACUAUGAAUCUCCAUA .......((((((((((.....((((((.(((..(((..(((....)))..))).))).))))))........))))).......))))).. ( -22.73, z-score = -0.91, R) >droYak2.chr3R 13412281 92 - 28832112 AUCAAUCGGAGGGUGCUCCUCUGUGGCAGUGCUUUCCCUCAUUUAAAUGCAGGACGCAUUGCUAUUUCACUAUGGCACUAUGAAUCUCCAUA .......((((((((((.....((((((((((..(((..(((....)))..))).))))))))))........))))))......))))... ( -28.82, z-score = -2.48, R) >droSec1.super_0 8223976 92 + 21120651 AUCUAUCUGUGGGUGCUCCUCUGUGGCAGUGCUUUCCCUCAUUUAAAUGCAGGACGCAUUGCUAUAUCACUAUGGCACUAUGGAUCUCCAUA ...........((((((....(((((((((((..(((..(((....)))..))).))))))))))).......))))))((((....)))). ( -28.40, z-score = -2.23, R) >droSim1.chr3R 15199486 92 + 27517382 AUCUAUCUGUGGGUGCUCCUCUGUGGCAGUGCUUUCCCUCAUUUAAAUGCAGGACGCAUUGCUAUUUCACUAUGGCACUAUGGAUCUCCAUA ...........((((((.....((((((((((..(((..(((....)))..))).))))))))))........))))))((((....)))). ( -27.92, z-score = -2.28, R) >consensus AUCUAUCUGUGGGUGCUCCUCUGUGGCAGUGCUUUCCCUCAUUUAAAUGCAGGACGCAUUGCUAUUUCACUAUGGCACUAUGAAUCUCCAUA ...........((((((.....((((((((((..(((..(((....)))..))).))))))))))........))))))((((....)))). (-24.24 = -24.68 + 0.44)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:11:23 2011