
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,041,238 – 9,041,340 |
| Length | 102 |
| Max. P | 0.681839 |

| Location | 9,041,238 – 9,041,338 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 87.77 |
| Shannon entropy | 0.19310 |
| G+C content | 0.58849 |
| Mean single sequence MFE | -43.78 |
| Consensus MFE | -33.19 |
| Energy contribution | -33.83 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.681839 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
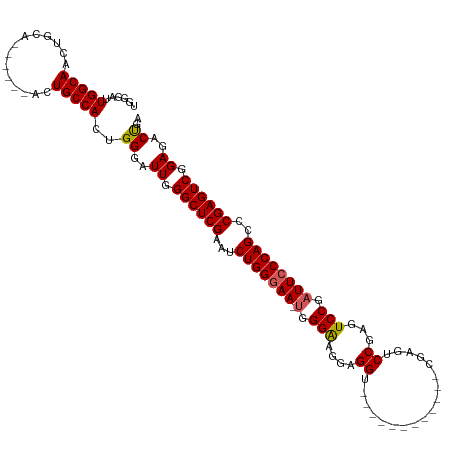
>dm3.chr3R 9041238 100 - 27905053 GGAUUGGCAACUGCA------ACUGCCACUGGGAUUGGGCUCGAAUCUGGGAA-UGGGAAGGAGGU------------CGAGUCCGAGUCCGAUUCCCAGCCCGAGUCGGAGACUGAGA ....(((((......------..)))))((.((.((.((((((...(((((((-(.(((.(((...------------....)))...))).))))))))..)))))).))..)).)). ( -42.00, z-score = -1.90, R) >droSim1.chr3R 15131317 100 + 27517382 GGAUUGGCAACAGCA------ACUGCCACUGGGAUUGGGCUCGAAUCUGGGAA-UGGGAAGGAGGU------------CGAAUCCGAGUCCGAUUCCCAGCCCGAGUCGGAGACUGAGA ....(((((......------..)))))((.((.((.((((((...(((((((-(.(((.(((...------------....)))...))).))))))))..)))))).))..)).)). ( -42.90, z-score = -2.52, R) >droSec1.super_0 8157154 100 + 21120651 GGAUUGGCAACAGCA------ACUGCCACUGGGAUUGGGCUCGAAUCUGGGAA-UGGGAAGGAGGU------------CGAAUCCGAGUCCGAUUCCCAGCCCGAGUCGGAGACUGAGA ....(((((......------..)))))((.((.((.((((((...(((((((-(.(((.(((...------------....)))...))).))))))))..)))))).))..)).)). ( -42.90, z-score = -2.52, R) >droYak2.chr3R 13342527 111 - 28832112 GGAUUGGCAACUGCAACUGCCACUGCCACUGGGAUUGGGCUCGAAUCUGGGAA--UGGGAGGAGGUCGAGA------CCGAGUCCGAGUCCGAUUCCCAGCCCGAGUCGGAGACCGAGA ((..(((((........)))))...)).((((..((.((((((...(((((((--(.(((((((((....)------))...)))...))).))))))))..)))))).))..)).)). ( -47.50, z-score = -1.73, R) >droEre2.scaffold_4770 5137925 119 + 17746568 GGAUUGGCAACUGCAACUGCCACUGCCACUGGGAUUGGGCUCGAAUCUGGAAAAUGGGAAGAAGGUCGAGACCGAGUCCGAGUCCGAGUCCGAUUCCCAGCCCGAGUCGGAGACUGAGA ((..(((((...((....))...)))))(((((((((((((((.(..((((...(((...(.....)....)))..))))..).))))))))).)))))).)).((((...)))).... ( -43.60, z-score = -0.92, R) >consensus GGAUUGGCAACUGCA______ACUGCCACUGGGAUUGGGCUCGAAUCUGGGAA_UGGGAAGGAGGU____________CGAGUCCGAGUCCGAUUCCCAGCCCGAGUCGGAGACUGAGA ....(((((..............)))))((.((.((.((((((...(((((((.((((..(((...................)))...)))).)))))))..)))))).))..)).)). (-33.19 = -33.83 + 0.64)



| Location | 9,041,240 – 9,041,340 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 87.41 |
| Shannon entropy | 0.19916 |
| G+C content | 0.58849 |
| Mean single sequence MFE | -44.22 |
| Consensus MFE | -33.79 |
| Energy contribution | -33.83 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.677600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9041240 100 - 27905053 UGGGAUUGGCAACUGCA------ACUGCCACUGGGAUUGGGCUCGAAUCUGGGAA-UGGGAAGGAGGU------------CGAGUCCGAGUCCGAUUCCCAGCCCGAGUCGGAGACUGA ......(((((......------..)))))..((..((.((((((...(((((((-(.(((.(((...------------....)))...))).))))))))..)))))).))..)).. ( -41.20, z-score = -1.59, R) >droSim1.chr3R 15131319 100 + 27517382 AGGGAUUGGCAACAGCA------ACUGCCACUGGGAUUGGGCUCGAAUCUGGGAA-UGGGAAGGAGGU------------CGAAUCCGAGUCCGAUUCCCAGCCCGAGUCGGAGACUGA ......(((((......------..)))))..((..((.((((((...(((((((-(.(((.(((...------------....)))...))).))))))))..)))))).))..)).. ( -42.10, z-score = -2.09, R) >droSec1.super_0 8157156 100 + 21120651 UGGGAUUGGCAACAGCA------ACUGCCACUGGGAUUGGGCUCGAAUCUGGGAA-UGGGAAGGAGGU------------CGAAUCCGAGUCCGAUUCCCAGCCCGAGUCGGAGACUGA ......(((((......------..)))))..((..((.((((((...(((((((-(.(((.(((...------------....)))...))).))))))))..)))))).))..)).. ( -42.10, z-score = -2.18, R) >droYak2.chr3R 13342529 111 - 28832112 UGGGAUUGGCAACUGCAACUGCCACUGCCACUGGGAUUGGGCUCGAAUCUGGGAA--UGGGAGGAGGUCGAGA------CCGAGUCCGAGUCCGAUUCCCAGCCCGAGUCGGAGACCGA (((.(.(((((........))))).).))).(((..((.((((((...(((((((--(.(((((((((....)------))...)))...))).))))))))..)))))).))..))). ( -48.90, z-score = -1.97, R) >droEre2.scaffold_4770 5137927 119 + 17746568 UGGGAUUGGCAACUGCAACUGCCACUGCCACUGGGAUUGGGCUCGAAUCUGGAAAAUGGGAAGAAGGUCGAGACCGAGUCCGAGUCCGAGUCCGAUUCCCAGCCCGAGUCGGAGACUGA .(((..(((((...((....))...)))))(((((((((((((((.(..((((...(((...(.....)....)))..))))..).))))))))).))))))))).((((...)))).. ( -46.80, z-score = -1.61, R) >consensus UGGGAUUGGCAACUGCA______ACUGCCACUGGGAUUGGGCUCGAAUCUGGGAA_UGGGAAGGAGGU____________CGAGUCCGAGUCCGAUUCCCAGCCCGAGUCGGAGACUGA .(((..(((((..............)))))((((((((((((((...(((........))).(((...................))))))))))).))))))))).....(....)... (-33.79 = -33.83 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:11:17 2011