
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,035,947 – 9,036,060 |
| Length | 113 |
| Max. P | 0.765127 |

| Location | 9,035,947 – 9,036,055 |
|---|---|
| Length | 108 |
| Sequences | 13 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.31 |
| Shannon entropy | 0.48358 |
| G+C content | 0.53479 |
| Mean single sequence MFE | -35.31 |
| Consensus MFE | -19.15 |
| Energy contribution | -18.80 |
| Covariance contribution | -0.35 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.616482 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9035947 108 - 27905053 ACAGAUCGCUUUACGAUGGGCCAGAAUCUGGCCAUCAUCUGGAGUACGGCGCCUUUGGAUGCCGAUGACGGUGAUUUCCCCUCUCGCAUCCAGAGUUGGUUUAACGAG-- ..(((((((((((.((((((((((...))))))..)))))))))).......((((((((((.((.((.((.(....))).))))))))))))))..)))))......-- ( -39.10, z-score = -2.06, R) >droAna3.scaffold_13340 1586255 108 - 23697760 AUAGAUCGCUUCACGAUGGGCCAGAAUCUGGCCAUUAUCUGGAGCACGGCGCCGUUGGAUGCCGAUGAUGGGGACUUCCCCUCGCGCAUCCAGAGUUGGUUCAACGAG-- ...((.(((((((.((((((((((...))))))..)))))))))).((((....(((((((((((....((((....))))))).)))))))).))))).))......-- ( -46.80, z-score = -2.95, R) >droEre2.scaffold_4770 5132605 108 + 17746568 ACAGAUCGCUUUACAAUGGGCCAGAAUCUGGCCAUUAUCUGGAGUACGGUCCCCUUGGACGCCGAUGACGGUGAUUUCCCCUCUCGCAUCCAGAGUUGGUUUAACGAG-- .....(((.....((((.((((((...))))))....((((((...((((((....))))((((....))))............))..))))))))))......))).-- ( -33.10, z-score = -0.78, R) >droYak2.chr3R 13337138 108 - 28832112 ACAGAUCGCUUUACGAUGGGCCAGAAUCUGGCCAUCAUCUGGAGUACGGCGCCCUUGGACGCCGAUGACGGUGAUUUUCCCUCUCGCAUCCAGAGUUGGUUUAACGAG-- ..((((((((((..((((((((((...))))))((((.(((..((.(((((((...)).)))))))..)))))))...........)))).)))).))))))......-- ( -36.10, z-score = -1.19, R) >droSec1.super_0 8151890 108 + 21120651 ACAGAUCGCUUUACGAUGGGCCAAAAUCUGGCUAUCAUCUGGAGUACGGCGCCCUUGGACGCCGAUGACGGUGAUUUCCCCUCUCGCAUCCAGAGUUGGUUUAACGAG-- .....(((.(..(((((((.(((.....)))))))).((((((((.(((((((...)).)))))..((.((.(....))).))..)).))))))....))..).))).-- ( -32.70, z-score = -0.49, R) >droSim1.chr3R 15125997 108 + 27517382 ACAGAUCGCUUUACGAUGGGACAAAAUCUGGCCAUCAUCUGGAGUACGGCGCCCUUGGACGCCGAUGACGGUGAUUUCCCCUCUCGUAUCCAAAGUUGGUUUAACGAG-- ....((((.....))))((((...((((..(((.(((((.(.....)((((((...)).))))))))).)))))))))))..(((((..(((....)))....)))))-- ( -31.40, z-score = -0.47, R) >droWil1.scaffold_181108 1287330 108 + 4707319 CUAGAUCGCUUCACAAUGGGUCAGAAUUUAGCCAUCAUUUGGAGCACAGCACCACUGGAAGCCGAUGAUGGCGAUUUCCCGUCACGCAUUCAAAACUGGUUCAAUGAA-- .............((.((..((((......(((((((((.((....(((.....)))....)))))))))))(((.....)))............))))..)).))..-- ( -29.40, z-score = -1.11, R) >droPer1.super_0 9409845 108 - 11822988 ACAGAUCGCUUCACCAUGGGCCAGAACUUGGCCAUUAUUUGGAGCACGGCGCCGCUGGACGCCGACGACGGGGACUUCCCCUCCCGCAUCCAGAGCUGGUUCAACGAG-- .....(((....((((..((((((...))))))....((((((((.(((((((...)).)))))..((.((((....))))))..)).))))))..))))....))).-- ( -41.70, z-score = -2.12, R) >dp4.chr2 19734318 108 + 30794189 ACAGAUCGCUUCACCAUGGGCCAGAACUUGGCCAUUAUCUGGAGCACGGCGCCGCUGGACGCCGACGACGGGGACUUCCCCUCCCGCAUCCAGAGCUGGUUCAACGAG-- .....(((....((((..((((((...))))))....((((((((.(((((((...)).)))))..((.((((....))))))..)).))))))..))))....))).-- ( -44.30, z-score = -2.64, R) >droGri2.scaffold_14906 7802088 108 + 14172833 ACAGAUCGCUUUACCAUGGGUCAAAAUCUGGCCGUUAUUUGGAGCACUGCACCGCUGGAAGCUAACGAUGGUGAUUUUCCAUCGCGCAUCCAGAACUGGUUCAAUGAG-- .....((((((((.....(((((.....)))))......))))))..((.(((.(((((.((...((((((.......)))))).)).)))))....))).))..)).-- ( -32.00, z-score = -1.17, R) >droMoj3.scaffold_6540 26127377 108 + 34148556 AUAGAUCGCUUUACCAUGGGCCAGAAUCUGGCUGUCAUCUGGAGCACUGCGCCACUGGACCCCGACGAUGGCGAUUUCCCAUCGCGCAUCCAGAACUGGUUCAAUGAG-- .....((((((((..(((((((((...))))))..))).))))))..((.(((((((((...((.((((((.(....)))))))))..)))))...)))).))..)).-- ( -35.40, z-score = -1.17, R) >droVir3.scaffold_12855 6434073 108 + 10161210 AUAGAUCGUUUUACCAUGGGCCAAAAUCUGGCUGUUAUAUGGAGCACUGCACCGCUAGAAGCGGAAGAUGGGGACUUUCCAUCGCGCAUUCAGAACUGGUUCAAUGAG-- .....(((((..((((..(((((.....)))))(((...((((((...((.((((.....))))..((((((.....)))))))))).)))).)))))))..))))).-- ( -34.50, z-score = -1.93, R) >triCas2.ChLG2 4268075 104 - 12900155 UCAGAACGUUUCAGUGUUGGUCAGAAUUUAUACUUGUGGUCCUCCACUGCACCGGAAAAGGGGUUCAAUG--CAACCAGUGCCGUGUAUUC----CUGGUUUAACGAACA ......((((.(((((..(((((.((.......)).))).))..)))))....(((..((((((.((..(--((.....)))..)).))))----))..))))))).... ( -22.50, z-score = 1.21, R) >consensus ACAGAUCGCUUUACCAUGGGCCAGAAUCUGGCCAUCAUCUGGAGCACGGCGCCGCUGGACGCCGAUGACGGUGAUUUCCCCUCGCGCAUCCAGAGCUGGUUCAACGAG__ .......((((((.....(((((.....)))))......))))))...(.(((.(((((.((....((.((.......)).))..)).)))))....))).)........ (-19.15 = -18.80 + -0.35)



| Location | 9,035,959 – 9,036,060 |
|---|---|
| Length | 101 |
| Sequences | 13 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 73.75 |
| Shannon entropy | 0.56100 |
| G+C content | 0.53356 |
| Mean single sequence MFE | -31.80 |
| Consensus MFE | -17.50 |
| Energy contribution | -17.41 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.765127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9035959 101 - 27905053 -----------UAUUAACAGAUCGCUUUACGAUGGGCCAGAAUCUGGCCAUCAUCUGGAGUACGGCGCCUUUGGAUGCCGAUGACGGUGAUUUCCCCUCUCGCAUCCAGAGU- -----------...........((((.(((((((((((((...))))))..))))....))).)))).((((((((((.((.((.((.(....))).)))))))))))))).- ( -37.10, z-score = -2.38, R) >droAna3.scaffold_13340 1586267 101 - 23697760 -----------CCUUAAUAGAUCGCUUCACGAUGGGCCAGAAUCUGGCCAUUAUCUGGAGCACGGCGCCGUUGGAUGCCGAUGAUGGGGACUUCCCCUCGCGCAUCCAGAGU- -----------............((((((.((((((((((...))))))..))))))))))(((....)))((((((((((....((((....))))))).)))))))....- ( -43.70, z-score = -2.88, R) >droEre2.scaffold_4770 5132617 101 + 17746568 -----------UACUGACAGAUCGCUUUACAAUGGGCCAGAAUCUGGCCAUUAUCUGGAGUACGGUCCCCUUGGACGCCGAUGACGGUGAUUUCCCCUCUCGCAUCCAGAGU- -----------.......................((((((...))))))....((((((...((((((....))))((((....))))............))..))))))..- ( -29.40, z-score = -0.34, R) >droYak2.chr3R 13337150 101 - 28832112 -----------UAUUGACAGAUCGCUUUACGAUGGGCCAGAAUCUGGCCAUCAUCUGGAGUACGGCGCCCUUGGACGCCGAUGACGGUGAUUUUCCCUCUCGCAUCCAGAGU- -----------...(((...((((.....)))).((((((...)))))).)))((((((((.(((((((...)).)))))..((.((.......)).))..)).))))))..- ( -34.00, z-score = -1.17, R) >droSec1.super_0 8151902 101 + 21120651 -----------UAUUAACAGAUCGCUUUACGAUGGGCCAAAAUCUGGCUAUCAUCUGGAGUACGGCGCCCUUGGACGCCGAUGACGGUGAUUUCCCCUCUCGCAUCCAGAGU- -----------...................(((((.(((.....)))))))).((((((((.(((((((...)).)))))..((.((.(....))).))..)).))))))..- ( -30.20, z-score = -0.74, R) >droSim1.chr3R 15126009 101 + 27517382 -----------UAUUAACAGAUCGCUUUACGAUGGGACAAAAUCUGGCCAUCAUCUGGAGUACGGCGCCCUUGGACGCCGAUGACGGUGAUUUCCCCUCUCGUAUCCAAAGU- -----------............(((((((((.(((...(((((..(((.(((((.(.....)((((((...)).))))))))).))))))))..))).)))))....))))- ( -28.10, z-score = -0.57, R) >droWil1.scaffold_181108 1287342 108 + 4707319 ----UGUUUCUGCUCACUAGAUCGCUUCACAAUGGGUCAGAAUUUAGCCAUCAUUUGGAGCACAGCACCACUGGAAGCCGAUGAUGGCGAUUUCCCGUCACGCAUUCAAAAC- ----...(((((((((...((.....))....)))).)))))....(((((((((.((....(((.....)))....)))))))))))(((.....))).............- ( -27.10, z-score = -0.61, R) >droPer1.super_0 9409857 101 - 11822988 -----------UGCUGACAGAUCGCUUCACCAUGGGCCAGAACUUGGCCAUUAUUUGGAGCACGGCGCCGCUGGACGCCGACGACGGGGACUUCCCCUCCCGCAUCCAGAGC- -----------(((.......((((((((.....((((((...))))))......)))))).(((((((...)).)))))..)).((((....))))....)))........- ( -37.00, z-score = -1.31, R) >dp4.chr2 19734330 101 + 30794189 -----------UGCUGACAGAUCGCUUCACCAUGGGCCAGAACUUGGCCAUUAUCUGGAGCACGGCGCCGCUGGACGCCGACGACGGGGACUUCCCCUCCCGCAUCCAGAGC- -----------((.(((.((....))))).))..((((((...))))))....((((((((.(((((((...)).)))))..((.((((....))))))..)).))))))..- ( -39.20, z-score = -1.70, R) >droGri2.scaffold_14906 7802100 112 + 14172833 UUUUUAAUGUUAACAAACAGAUCGCUUUACCAUGGGUCAAAAUCUGGCCGUUAUUUGGAGCACUGCACCGCUGGAAGCUAACGAUGGUGAUUUUCCAUCGCGCAUCCAGAAC- .......((((....))))....((((((.....(((((.....)))))......)))))).........(((((.((...((((((.......)))))).)).)))))...- ( -29.20, z-score = -1.19, R) >droMoj3.scaffold_6540 26127389 101 + 34148556 -----------UACCCAUAGAUCGCUUUACCAUGGGCCAGAAUCUGGCUGUCAUCUGGAGCACUGCGCCACUGGACCCCGACGAUGGCGAUUUCCCAUCGCGCAUCCAGAAC- -----------............((((((..(((((((((...))))))..))).)))))).........(((((...((.((((((.(....)))))))))..)))))...- ( -31.30, z-score = -1.25, R) >droVir3.scaffold_12855 6434085 109 + 10161210 ---UGCCCUCCCUUUAAUAGAUCGUUUUACCAUGGGCCAAAAUCUGGCUGUUAUAUGGAGCACUGCACCGCUAGAAGCGGAAGAUGGGGACUUUCCAUCGCGCAUUCAGAAC- ---(((.((((...(((((..((((......))))((((.....)))))))))...))))....((.((((.....))))..((((((.....)))))))))))........- ( -29.50, z-score = -0.26, R) >triCas2.ChLG2 4268098 86 - 12900155 -----------UAUUUUCAGAACGUUUCAGUGUUGGUCAGAAUUUA-----UACUUGUGGUCCUCCACUGC-----ACCGGAAAAGGGG---UUCAAU---GCAACCAGUGCC -----------........((((....(((((..(((((.((....-----...)).))).))..))))).-----.((......)).)---)))...---(((.....))). ( -17.60, z-score = 0.77, R) >consensus ___________UAUUAACAGAUCGCUUUACCAUGGGCCAGAAUCUGGCCAUUAUCUGGAGCACGGCGCCGCUGGACGCCGAUGACGGUGAUUUCCCCUCUCGCAUCCAGAGC_ .......................((((((.....(((((.....)))))......)))))).........(((((.((....((.((.......)).))..)).))))).... (-17.50 = -17.41 + -0.10)

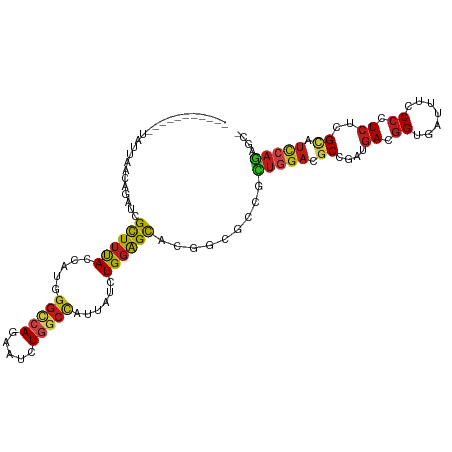
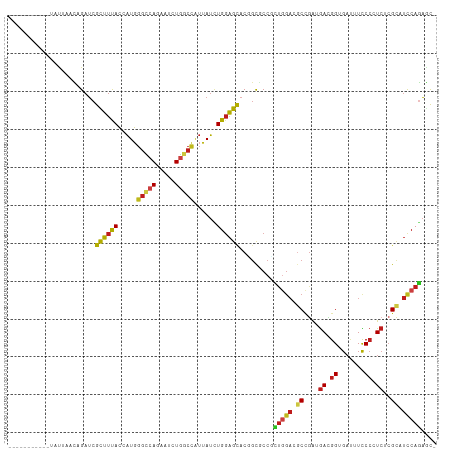
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:11:15 2011