| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,023,104 – 9,023,198 |
| Length | 94 |
| Max. P | 0.733966 |

| Location | 9,023,104 – 9,023,198 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 74.52 |
| Shannon entropy | 0.51763 |
| G+C content | 0.49688 |
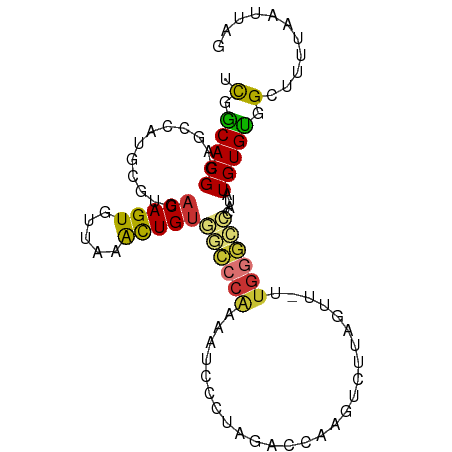
| Mean single sequence MFE | -27.75 |
| Consensus MFE | -12.04 |
| Energy contribution | -11.44 |
| Covariance contribution | -0.59 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.733966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
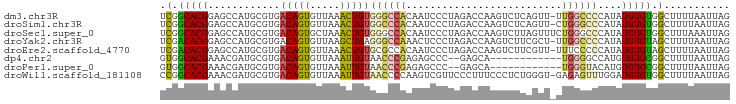
>dm3.chr3R 9023104 94 + 27905053 UCGGCACGGAGCCAUGCGUGACAGUGUUAAACUGUGGGCCACAAUCCCUAGACCAAGUCUCAGUU-UUGGCCCCAUAUGUGUGGCUUUUAAUUAG .......((((((((((((((((((.....)))))((((((.(((....((((...))))..)))-.))))))..)))))))))))))....... ( -34.40, z-score = -2.97, R) >droSim1.chr3R 15113194 94 - 27517382 UCGGCACGGAGCCAUGCGUGACAGUGUUAAACUGUGGCCCACAAUCCCUAGACCAAGUCUCAGUU-CUGGGCCCAUAUGUGUGGCUUUUAAUUAG .......((((((((((((((((((.....)))))((((((.(((....((((...))))..)))-.))))))..)))))))))))))....... ( -34.40, z-score = -2.85, R) >droSec1.super_0 8139122 95 - 21120651 UCGGCACGGAGCCAUGCGUGACAGUGCUAAACUGUGGGCCACAAUCCCUAGACCAAGUCUUAGUUUCUGGGCCCAUAUGUGUGGCUUUAAAUUAG .......((((((((((((((((((.....)))))(((((.((....((((((...))).)))....))))))).)))))))))))))....... ( -35.80, z-score = -2.91, R) >droYak2.chr3R 13324178 94 + 28832112 UCGACACGGAGCCAUGCGUGACAGUGUUAAGCUGAGGGCCAAACUCCCUAGACCAAGUCUUCGCU-UUGGCCCCAUAUGUGUAGCUUUUAAUUAG .......(((((.(((((((.((((.....)))).((((((((......((((...))))....)-)))))))..))))))).)))))....... ( -28.00, z-score = -1.73, R) >droEre2.scaffold_4770 5119817 94 - 17746568 UCGACACGGAGCCAUGCGUGACAGUGUUAAACUGUGCGCCACAAUCCCUAGACCAAGUCUUCGUU-UUUCCCCCAUAUGUGUAGCUUUUAAUUAG ...(((((((.....((((.(((((.....))))))))).....)))..((((...)))).....-............))))............. ( -19.60, z-score = -1.29, R) >dp4.chr2 19720939 81 - 30794189 GUGGCACGAAACGAUGCGUGACAGUGUUAAAUUGUAACCCGAGAGCCC--GAGCA------------UGGGGCCAUGUGUGCGGCUUUUAAUUAG ..((((((........))))(((((.....)))))...))((((((((--(.(((------------((....))))).)).)))))))...... ( -24.30, z-score = -0.98, R) >droPer1.super_0 9396003 81 + 11822988 GUGGCACGAAACGAUGCGUGACAGUGUUAAAUUGUAACCCGAGAGCCC--GAGCA------------UGGGUACAUGUGUGCGGCUUUUAAUUAG ..((((((........))))(((((.....)))))...))((((((((--(.(((------------((....))))).)).)))))))...... ( -23.80, z-score = -1.24, R) >droWil1.scaffold_181108 1274822 94 - 4707319 CCGGCACGAAACGAUGCGUGACAGUGUUAAAUUGUAACCCCAAGUCGUUCCCUUUCCCUCUGGGU-GAGAGUUUGGAUGUGUGGCUUUUAAUUAG .(.(((((..(((((..((.(((((.....))))).)).....)))))((((((((((....)).-)))))...)))))))).)........... ( -21.70, z-score = -0.07, R) >consensus UCGGCACGGAGCCAUGCGUGACAGUGUUAAACUGUGGCCCAAAAUCCCUAGACCAAGUCUUAGUU_UUGGGCCCAUAUGUGUGGCUUUUAAUUAG .(.(((((............(((((.....)))))((((((..........................))))))....))))).)........... (-12.04 = -11.44 + -0.59)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:11:13 2011