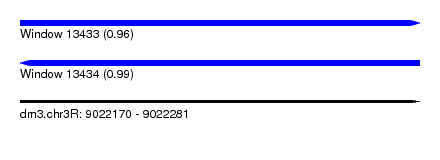
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,022,170 – 9,022,281 |
| Length | 111 |
| Max. P | 0.993316 |

| Location | 9,022,170 – 9,022,281 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 90.90 |
| Shannon entropy | 0.16015 |
| G+C content | 0.37366 |
| Mean single sequence MFE | -29.48 |
| Consensus MFE | -26.66 |
| Energy contribution | -27.98 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.960749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
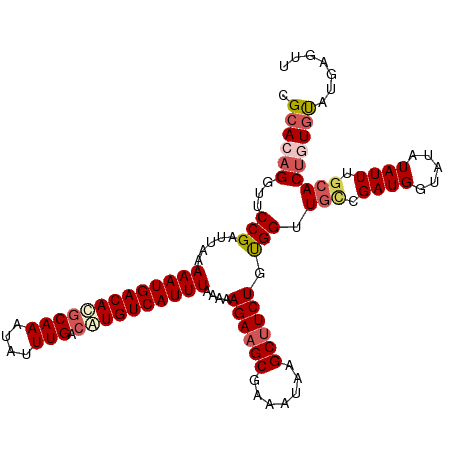
>dm3.chr3R 9022170 111 + 27905053 CGCACAGGUUCCGAUUAAAAAUGACAUGCAAAUAUUUGACAUGUCAUUUAAAAAGAAGCGAAAUAAGCUUCUGUGGUUGUUGAUGGUAUAUAUUUGCACUGUGUAUGAGUU .((((((....((((((.(((((((((((((....))).))))))))))....((((((.......)))))).))))))......(((......))).))))))....... ( -32.40, z-score = -3.43, R) >droEre2.scaffold_4770 5118855 110 - 17746568 AGCACAGAUUCCGAUUAAAAAUGACACGCAAGUGCUUGAAGUGUCAUUUAAAAAGAAGCGAA-UAAGCUUCUGUGGCUAUCGAUGGUAUAUAUUCCCACCAUGUAUGAGUU ......(((((((((...(((((((((.(((....)))..)))))))))....((((((...-...))))))......))))(((((..........)))))....))))) ( -26.80, z-score = -1.89, R) >droYak2.chr3R 13322280 111 + 28832112 UGCACAGGUUCCGAUUAAAAAUGACACGCAAAUAUUUGGCGUGUCAUUUAAAAAGAAGCGAAAUAAGCUUCUGUGGUUGCCGAUGGUAUAUAUUCGCACUAUGCAUGAGUU ((((.((...(((.....(((((((((((.........)))))))))))....((((((.......)))))).))).(((.((((.....)))).))))).))))...... ( -32.80, z-score = -2.64, R) >droSec1.super_0 8138177 111 - 21120651 CGCACAGGUUCCGAUUAAAAAUGACACGCAAAUAUUUGACAUGUCAUUUAAAAAGAAGCGAAAUAAGCUUCUGUGGUUGCCGAUGGUAUAUAUUUGCACUGUGCAUGAGUU .((((((...(((.....((((((((.((((....))).).))))))))....((((((.......)))))).))).(((.((((.....)))).)))))))))....... ( -31.80, z-score = -2.67, R) >droSim1.chr3R 15112237 111 - 27517382 CGCACAGGUUCCGAUUAAAAAUGACACGCAAAUAUUUGACAUGUCAUUUAAAAAGAAGCGAAAUAAGCGUCUGCGGUUGCCGAUGGUAUAUAUUUGCACUGUAUAUGAGUU ..((..(((.(((.....((((((((.((((....))).).))))))))....(((.((.......)).))).)))..)))..)).(((((((.......))))))).... ( -23.60, z-score = -0.08, R) >consensus CGCACAGGUUCCGAUUAAAAAUGACACGCAAAUAUUUGACAUGUCAUUUAAAAAGAAGCGAAAUAAGCUUCUGUGGUUGCCGAUGGUAUAUAUUUGCACUGUGUAUGAGUU .((((((...(((.....(((((((((((((....))).))))))))))....((((((.......)))))).))).(((.((((.....)))).)))))))))....... (-26.66 = -27.98 + 1.32)

| Location | 9,022,170 – 9,022,281 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 90.90 |
| Shannon entropy | 0.16015 |
| G+C content | 0.37366 |
| Mean single sequence MFE | -27.46 |
| Consensus MFE | -22.52 |
| Energy contribution | -22.20 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.993316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
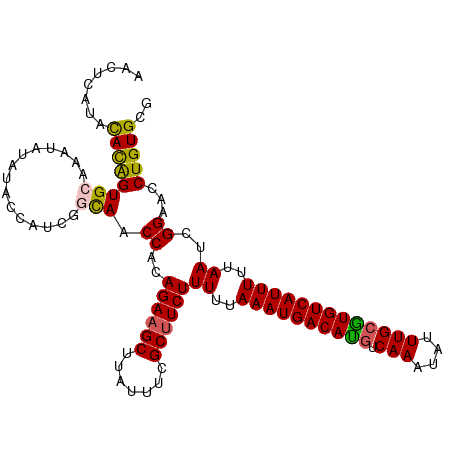
>dm3.chr3R 9022170 111 - 27905053 AACUCAUACACAGUGCAAAUAUAUACCAUCAACAACCACAGAAGCUUAUUUCGCUUCUUUUUAAAUGACAUGUCAAAUAUUUGCAUGUCAUUUUUAAUCGGAACCUGUGCG ........(((((..........................((((((.......))))))((..((((((((((.(((....)))))))))))))..)).......))))).. ( -24.30, z-score = -3.75, R) >droEre2.scaffold_4770 5118855 110 + 17746568 AACUCAUACAUGGUGGGAAUAUAUACCAUCGAUAGCCACAGAAGCUUA-UUCGCUUCUUUUUAAAUGACACUUCAAGCACUUGCGUGUCAUUUUUAAUCGGAAUCUGUGCU ......((((.(((((.........)))))(((..((..((((((...-...))))))((..(((((((((..(((....))).)))))))))..))..)).))))))).. ( -27.70, z-score = -2.69, R) >droYak2.chr3R 13322280 111 - 28832112 AACUCAUGCAUAGUGCGAAUAUAUACCAUCGGCAACCACAGAAGCUUAUUUCGCUUCUUUUUAAAUGACACGCCAAAUAUUUGCGUGUCAUUUUUAAUCGGAACCUGUGCA ......((((((((((((..........)).))).((..((((((.......))))))((..(((((((((((.........)))))))))))..))..))...))))))) ( -31.80, z-score = -4.15, R) >droSec1.super_0 8138177 111 + 21120651 AACUCAUGCACAGUGCAAAUAUAUACCAUCGGCAACCACAGAAGCUUAUUUCGCUUCUUUUUAAAUGACAUGUCAAAUAUUUGCGUGUCAUUUUUAAUCGGAACCUGUGCG .......(((((((((..((.......))..))).((..((((((.......))))))((..((((((((((.(((....)))))))))))))..))..))...)))))). ( -31.30, z-score = -4.33, R) >droSim1.chr3R 15112237 111 + 27517382 AACUCAUAUACAGUGCAAAUAUAUACCAUCGGCAACCGCAGACGCUUAUUUCGCUUCUUUUUAAAUGACAUGUCAAAUAUUUGCGUGUCAUUUUUAAUCGGAACCUGUGCG ........((((((((..((.......))..))).(((.(((.((.......)).)))((..((((((((((.(((....)))))))))))))..)).)))...))))).. ( -22.20, z-score = -1.25, R) >consensus AACUCAUACACAGUGCAAAUAUAUACCAUCGGCAACCACAGAAGCUUAUUUCGCUUCUUUUUAAAUGACAUGUCAAAUAUUUGCGUGUCAUUUUUAAUCGGAACCUGUGCG ........((((((((...............))).((..((((((.......))))))((..((((((((((.(((....)))))))))))))..))..))...))))).. (-22.52 = -22.20 + -0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:11:13 2011