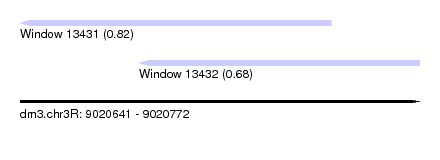
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 9,020,641 – 9,020,772 |
| Length | 131 |
| Max. P | 0.818797 |

| Location | 9,020,641 – 9,020,743 |
|---|---|
| Length | 102 |
| Sequences | 9 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 82.22 |
| Shannon entropy | 0.33781 |
| G+C content | 0.51730 |
| Mean single sequence MFE | -32.90 |
| Consensus MFE | -19.80 |
| Energy contribution | -19.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.818797 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
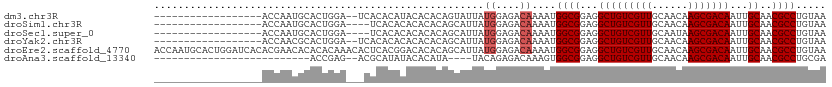
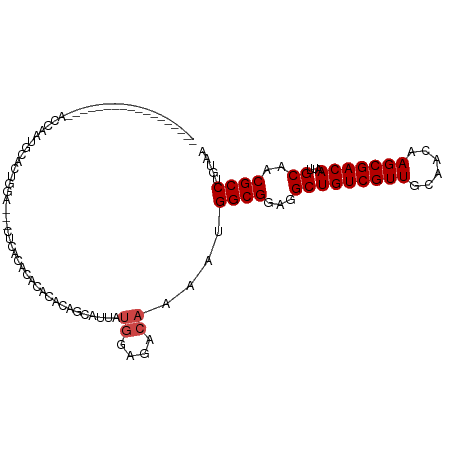
>dm3.chr3R 9020641 102 - 27905053 ---------------GUAUUAUGGAGACAAAAUGGCGGAGGCUGUCGUUGCAACAAGCGACAAUUGCAACGCCUGUAAGCCCGAAAGAGCGGAAACGGAAACGUGCAUAGUGCAUGC ---------------((((((((..........((((...(((((((((......)))))))...))..)))).....((.(....).))....(((....))).)))))))).... ( -35.80, z-score = -2.90, R) >droSim1.chr3R 15110721 102 + 27517382 ---------------GCAUUAUGGAGACAAAAUGGCGGAGGCUGUCGUUGCAACAAGCGACAAUUGCAACGCCUGUAAGCCCGAAAGAGCGGAAACGGAAACGUGCAUAGUGCAUGC ---------------((((((((..........((((...(((((((((......)))))))...))..)))).....((.(....).))....(((....))).)))))))).... ( -37.80, z-score = -3.21, R) >droSec1.super_0 8136657 102 + 21120651 ---------------GCAUUAUGGAGACAAAAUGGCGGAGGCUGUCGUUGCAAUAAGCGACAAUUGCAACGCCUGUAAGCCCGAAAGAGCGGAAACGGAAACGUGCAUAGUGCAUGC ---------------((((((((..........((((...(((((((((......)))))))...))..)))).....((.(....).))....(((....))).)))))))).... ( -37.80, z-score = -3.45, R) >droYak2.chr3R 13320713 102 - 28832112 ---------------GCAUUAUGGAGACAAAAUGGCGGAGGCUGUCGUUGCAACAAGCGACAAUUGCAACGCCUGUAAGCCCGAAAGAGCGGAAACGGAAACGUGCAUAGUGCAUGC ---------------((((((((..........((((...(((((((((......)))))))...))..)))).....((.(....).))....(((....))).)))))))).... ( -37.80, z-score = -3.21, R) >droEre2.scaffold_4770 5117262 117 + 17746568 ACUCACGGACACACAGCAUUAUGGAGACAAAAUGGCGGAGGCUGUCGUUGCAACAAGCGACAAUUGCAACGCCUGUAAGCCCGAAAGAGCGGAAACGGAAACGUGCAUAGUGCAUGC ...............((((((((..........((((...(((((((((......)))))))...))..)))).....((.(....).))....(((....))).)))))))).... ( -38.00, z-score = -2.05, R) >droAna3.scaffold_13340 1571024 98 - 23697760 -------------------UACAGAGACAAAGUGGCGGAGGCUGUCGUUGCAACAAGCGACAAUUGCAACGCCUGCGAAGCCCGAAAAGCGGAAACGGAAACGUGCAUAGUGCAUGC -------------------..............((((...(((((((((......)))))))...))..))))((((..(((((.....)))..(((....)))))....))))... ( -29.90, z-score = -0.65, R) >dp4.chr2 19716542 101 + 30794189 --------------GACAAGAGAGAAGAGAUACAGAGGACGUUGCCAUUGCAAUAAGCGACAAUUGCAACGCCUGCAAGGCCCAA--AGCGGAAACGGAAACGUGCAUAGUGCAUGC --------------.....................(((.((((((.(((((.......).)))).)))))))))(((..((((..--...))..(((....)))))....))).... ( -25.60, z-score = -1.57, R) >droPer1.super_0 9391555 101 - 11822988 --------------GACAAGAGAGAAGAGAUACAGAGGACGUUGCCAUUGCAAUAAGCGACAAUUGCAACGCCUGCAAGGCCCAA--AGCGGAAACGGAAACGUGCAUAGUGCAUGC --------------.....................(((.((((((.(((((.......).)))).)))))))))(((..((((..--...))..(((....)))))....))).... ( -25.60, z-score = -1.57, R) >droWil1.scaffold_181108 1272356 101 + 4707319 --------------GAGACGACUGAGGCGGAGAUUUGGA--CUGUCGUUGCAACAAGCGACAAUUGCAACGCCUGCAAAGCCCAAAAAGCGGAAACGGAAGUGUGCAUAGUGCAUGC --------------........((.(((...........--.(((((((......))))))).(((((.....))))).))))).....((....))...((((((.....)))))) ( -27.80, z-score = -0.14, R) >consensus _______________GCAUUAUGGAGACAAAAUGGCGGAGGCUGUCGUUGCAACAAGCGACAAUUGCAACGCCUGUAAGCCCGAAAGAGCGGAAACGGAAACGUGCAUAGUGCAUGC ....................................((....(((((((......))))))).(((((.....)))))..)).......((....))....(((((.....))))). (-19.80 = -19.80 + 0.00)


| Location | 9,020,680 – 9,020,772 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 80.24 |
| Shannon entropy | 0.34068 |
| G+C content | 0.48960 |
| Mean single sequence MFE | -24.57 |
| Consensus MFE | -18.83 |
| Energy contribution | -19.17 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.683245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 9020680 92 - 27905053 ------------------ACCAAUGCACUGGA--UCACACAUACACACAGUAUUAUGGAGACAAAAUGGCGGAGGCUGUCGUUGCAACAAGCGACAAUUGCAACGCCUGUAA ------------------.(((......))).--.......((((......(((.((....)).)))((((...(((((((((......)))))))...))..)))))))). ( -24.50, z-score = -1.23, R) >droSim1.chr3R 15110760 90 + 27517382 ------------------ACCAAUGCACUGGA----UCACACACACACAGCAUUAUGGAGACAAAAUGGCGGAGGCUGUCGUUGCAACAAGCGACAAUUGCAACGCCUGUAA ------------------.(((......))).----..........((((((((.((....)).))))(((...(((((((((......)))))))...))..))))))).. ( -25.30, z-score = -1.39, R) >droSec1.super_0 8136696 90 + 21120651 ------------------ACCAAUGCACUGGA----UCACACACACACAGCAUUAUGGAGACAAAAUGGCGGAGGCUGUCGUUGCAAUAAGCGACAAUUGCAACGCCUGUAA ------------------.(((......))).----..........((((((((.((....)).))))(((...(((((((((......)))))))...))..))))))).. ( -25.30, z-score = -1.49, R) >droYak2.chr3R 13320752 92 - 28832112 ------------------ACCAACGCACUGGA--UCACACACACACACAGCAUUAUGGAGACAAAAUGGCGGAGGCUGUCGUUGCAACAAGCGACAAUUGCAACGCCUGUAA ------------------.(((......))).--............((((((((.((....)).))))(((...(((((((((......)))))))...))..))))))).. ( -25.30, z-score = -1.52, R) >droEre2.scaffold_4770 5117301 112 + 17746568 ACCAAUGCACUGGAUCACACGAACACACACAAACACUCACGGACACACAGCAUUAUGGAGACAAAAUGGCGGAGGCUGUCGUUGCAACAAGCGACAAUUGCAACGCCUGUAA ...(((((..((..((....((..............))...))..))..))))).((....))....((((...(((((((((......)))))))...))..))))..... ( -25.34, z-score = -0.71, R) >droAna3.scaffold_13340 1571063 80 - 23697760 --------------------------ACCGAG--ACGCAUAUACACAUA----UACAGAGACAAAGUGGCGGAGGCUGUCGUUGCAACAAGCGACAAUUGCAACGCCUGCGA --------------------------......--.((((((((....))----))............((((...(((((((((......)))))))...))..)))))))). ( -21.70, z-score = -1.26, R) >consensus __________________ACCAAUGCACUGGA___CUCACACACACACAGCAUUAUGGAGACAAAAUGGCGGAGGCUGUCGUUGCAACAAGCGACAAUUGCAACGCCUGUAA .......................................................((....))....((((...(((((((((......)))))))...))..))))..... (-18.83 = -19.17 + 0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:11:11 2011