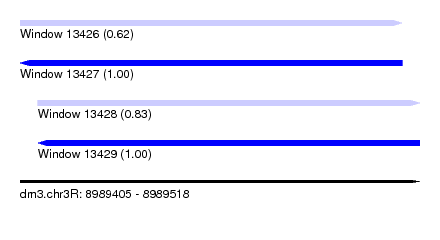
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,989,405 – 8,989,518 |
| Length | 113 |
| Max. P | 0.997915 |

| Location | 8,989,405 – 8,989,513 |
|---|---|
| Length | 108 |
| Sequences | 10 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.74 |
| Shannon entropy | 0.36272 |
| G+C content | 0.58557 |
| Mean single sequence MFE | -40.80 |
| Consensus MFE | -21.46 |
| Energy contribution | -22.02 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.617713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
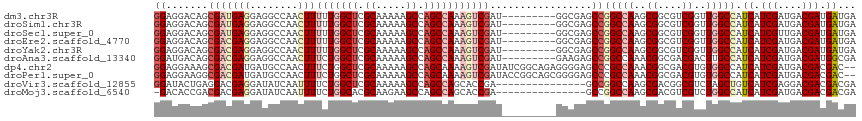
>dm3.chr3R 8989405 108 + 27905053 GGAGGACAGCGAUGAGGAGGCCAACUUUUUGGCUCGCAAAAAGCCAGCCAAAGUCGAU---------GGCGAGCCGGCCAAGCGGCGUCGGUUGGCCAUCAUCGAUGACGAUGAUGA ......((.((((((...((((((((.(((((((.((.....)).)))))))((((.(---------(((......))))..))))...)))))))).)))))).)).......... ( -45.60, z-score = -2.82, R) >droSim1.chr3R 15079428 108 - 27517382 GGAGGACAGCGAUGAGGAGGCCAACUUUUUGGCUCGCAAAAAGCCAGCCAAAGUCGAU---------GGCGAGCCGGCCAAGCGGCGUCGGUUGGCCAUCAUCGAUGACGAUGAUGA ......((.((((((...((((((((.(((((((.((.....)).)))))))((((.(---------(((......))))..))))...)))))))).)))))).)).......... ( -45.60, z-score = -2.82, R) >droSec1.super_0 8105532 108 - 21120651 GGAGGACAGCGAUGAGGAGGCCAACUUUUUGGCUCGCAAAAAGCCAGCCAAAGUCGAU---------GGCGAGCCGGCCAAGCGGCGUCGGUUGGCCAUCAUCGUUGACGAUGAUGA ......(((((((((...((((((((.(((((((.((.....)).)))))))((((.(---------(((......))))..))))...)))))))).))))))))).......... ( -49.80, z-score = -3.88, R) >droEre2.scaffold_4770 5085276 108 - 17746568 GGAGGACAGCGACGAGGAGGCCAACUUUUUGGCUCGCAAAAAGCCAGCCAAAGUCGAU---------GGCGAGCCGGCCAAGCGGCGUCGGUUGGCCAUCAUCGAUGACGAUGAUGA ........(((((((((((.....)))))))..)))).....(((((((...((((.(---------(((......))))..))))...)))))))((((((((....)))))))). ( -45.60, z-score = -2.74, R) >droYak2.chr3R 13284390 108 + 28832112 GGAGGACAGCGACGAGGAGGCCAACUUUUUGGCUCGCAAAAAGCCAGCCAAAGUCGAU---------GGCGAGCCGGCCAAGCGGCGUCGGUUGGCCAUCAUCGAUGACGAUGAUGA ........(((((((((((.....)))))))..)))).....(((((((...((((.(---------(((......))))..))))...)))))))((((((((....)))))))). ( -45.60, z-score = -2.74, R) >droAna3.scaffold_13340 1540200 108 + 23697760 GGAUGACAGCGACGAGGAGGCCAACUUUCUGGCUCGCAAAAAGCCAGCCAAAGUCGAU---------GAAGAGCCGGCCAAACGGCGACGACUUGCCAUCAUCGAUGACGAUGGCGA ......((.((((.((((((....))))))((((.((.....)).))))...)))).)---------)....((((......))))......((((((((.((...)).)))))))) ( -37.50, z-score = -1.82, R) >dp4.chr2 19689712 115 - 30794189 GGAGGAAAGCGACGAUGAUGCCAACUUUCUGGCUCGCAAAAAGCCAGCAAAAGUCGAUAUCGGCAGAGGGGAGCCCGCCAAACGGCGACGUGUGGCCAUCAUCGAUGACGACGAC-- (..(.....((.(((((((((((.((((((.(((.((.....)).)))....((((....)))))))))).....((((....)))).....))).)))))))).)).)..)...-- ( -38.40, z-score = -1.17, R) >droPer1.super_0 9365197 115 + 11822988 GGAGGAAGGCGACGAUGAUGCCAACUUUCUGGCUCGCAAAAAGCCAGCAAAAGUCGAUACCGGCAGCGGGGAGCCCGCCAAACGGCGACGUGUGGCCAUCAUCGAUGACGACGAC-- (..(.....((.((((((((((((((((((((((.......)))))).))).((((...(((...(((((...)))))....))))))))).))).)))))))).)).)..)...-- ( -41.30, z-score = -1.32, R) >droVir3.scaffold_12855 6375195 102 - 10161210 GGAUACUGAGGACGAGGAUAUCAAUUUUCUGGCUCGCAAAAAGCCAGCCAGCACCGA---------------GCCGGCCAAGCGACGGCGUCUAGCUGUCAUCGAGGACGACGACGA .........((((...............((((((.((.....)).))))))......---------------((((.........))))))))...((((.(((....))).)))). ( -28.90, z-score = 0.52, R) >droMoj3.scaffold_6540 26064244 101 - 34148556 -GACACCGACGACGAGGAUAUCAAUUUUCUGGCACGCAAGAAGCCAGCCAGCACCGA---------------GCCGGCCAAGCGACGUCGUCUGGCCAUCAUCGAUGACGACGACGA -..((.(((.((...((...........(((((.(....)..)))))..........---------------.))(((((..((....))..))))).)).))).)).......... ( -29.75, z-score = -0.77, R) >consensus GGAGGACAGCGACGAGGAGGCCAACUUUCUGGCUCGCAAAAAGCCAGCCAAAGUCGAU_________GGCGAGCCGGCCAAGCGGCGUCGGUUGGCCAUCAUCGAUGACGAUGACGA ............(((.((........((((((((.((.....)).))))))))......................(((((..((....))..))))).)).)))............. (-21.46 = -22.02 + 0.56)
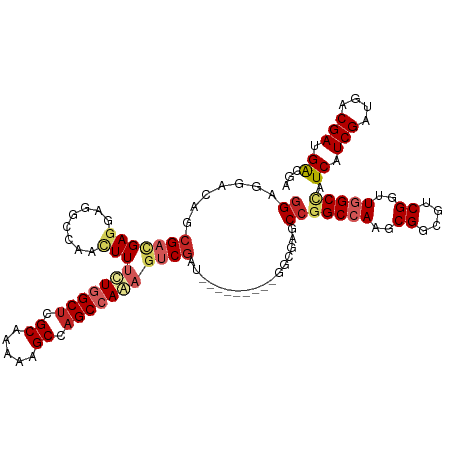
| Location | 8,989,405 – 8,989,513 |
|---|---|
| Length | 108 |
| Sequences | 10 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.74 |
| Shannon entropy | 0.36272 |
| G+C content | 0.58557 |
| Mean single sequence MFE | -43.18 |
| Consensus MFE | -32.18 |
| Energy contribution | -30.97 |
| Covariance contribution | -1.21 |
| Combinations/Pair | 1.42 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.21 |
| SVM RNA-class probability | 0.997915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8989405 108 - 27905053 UCAUCAUCGUCAUCGAUGAUGGCCAACCGACGCCGCUUGGCCGGCUCGCC---------AUCGACUUUGGCUGGCUUUUUGCGAGCCAAAAAGUUGGCCUCCUCAUCGCUGUCCUCC ........(.((.((((((.((((((((((.((((......)))))))..---------......(((((((.((.....)).)))))))..)))))))...)))))).)).).... ( -44.20, z-score = -3.90, R) >droSim1.chr3R 15079428 108 + 27517382 UCAUCAUCGUCAUCGAUGAUGGCCAACCGACGCCGCUUGGCCGGCUCGCC---------AUCGACUUUGGCUGGCUUUUUGCGAGCCAAAAAGUUGGCCUCCUCAUCGCUGUCCUCC ........(.((.((((((.((((((((((.((((......)))))))..---------......(((((((.((.....)).)))))))..)))))))...)))))).)).).... ( -44.20, z-score = -3.90, R) >droSec1.super_0 8105532 108 + 21120651 UCAUCAUCGUCAACGAUGAUGGCCAACCGACGCCGCUUGGCCGGCUCGCC---------AUCGACUUUGGCUGGCUUUUUGCGAGCCAAAAAGUUGGCCUCCUCAUCGCUGUCCUCC ........(.((.((((((.((((((((((.((((......)))))))..---------......(((((((.((.....)).)))))))..)))))))...)))))).)).).... ( -44.20, z-score = -3.99, R) >droEre2.scaffold_4770 5085276 108 + 17746568 UCAUCAUCGUCAUCGAUGAUGGCCAACCGACGCCGCUUGGCCGGCUCGCC---------AUCGACUUUGGCUGGCUUUUUGCGAGCCAAAAAGUUGGCCUCCUCGUCGCUGUCCUCC ........(.((.((((((.((((((((((.((((......)))))))..---------......(((((((.((.....)).)))))))..)))))))...)))))).)).).... ( -43.80, z-score = -3.48, R) >droYak2.chr3R 13284390 108 - 28832112 UCAUCAUCGUCAUCGAUGAUGGCCAACCGACGCCGCUUGGCCGGCUCGCC---------AUCGACUUUGGCUGGCUUUUUGCGAGCCAAAAAGUUGGCCUCCUCGUCGCUGUCCUCC ........(.((.((((((.((((((((((.((((......)))))))..---------......(((((((.((.....)).)))))))..)))))))...)))))).)).).... ( -43.80, z-score = -3.48, R) >droAna3.scaffold_13340 1540200 108 - 23697760 UCGCCAUCGUCAUCGAUGAUGGCAAGUCGUCGCCGUUUGGCCGGCUCUUC---------AUCGACUUUGGCUGGCUUUUUGCGAGCCAGAAAGUUGGCCUCCUCGUCGCUGUCAUCC ..(((((((((...)))))))))..(.((.((.((...(((((((((...---------...)).(((((((.((.....)).)))))))..)))))))....)).)).)).).... ( -43.50, z-score = -3.03, R) >dp4.chr2 19689712 115 + 30794189 --GUCGUCGUCAUCGAUGAUGGCCACACGUCGCCGUUUGGCGGGCUCCCCUCUGCCGAUAUCGACUUUUGCUGGCUUUUUGCGAGCCAGAAAGUUGGCAUCAUCGUCGCUUUCCUCC --(((((((....)))))))(((.((....((((....))))(((........)))(((..((((((...(((((((.....))))))).))))))..)))...)).)))....... ( -42.20, z-score = -2.29, R) >droPer1.super_0 9365197 115 - 11822988 --GUCGUCGUCAUCGAUGAUGGCCACACGUCGCCGUUUGGCGGGCUCCCCGCUGCCGGUAUCGACUUUUGCUGGCUUUUUGCGAGCCAGAAAGUUGGCAUCAUCGUCGCCUUCCUCC --(((((((....)))))))(((.((.((..((((...((((((...))))))..))))..)).....(((..((((((((.....))))))))..))).....)).)))....... ( -45.60, z-score = -2.45, R) >droVir3.scaffold_12855 6375195 102 + 10161210 UCGUCGUCGUCCUCGAUGACAGCUAGACGCCGUCGCUUGGCCGGC---------------UCGGUGCUGGCUGGCUUUUUGCGAGCCAGAAAAUUGAUAUCCUCGUCCUCAGUAUCC ..(((((((....))))))).(((.(((((((((....)).))))---------------(((((.((((((.((.....)).))))))...))))).......)))...))).... ( -38.60, z-score = -2.10, R) >droMoj3.scaffold_6540 26064244 101 + 34148556 UCGUCGUCGUCAUCGAUGAUGGCCAGACGACGUCGCUUGGCCGGC---------------UCGGUGCUGGCUGGCUUCUUGCGUGCCAGAAAAUUGAUAUCCUCGUCGUCGGUGUC- .((((((((....))))))))(((.(((((((......(((((((---------------.....)))))))(((.........)))................))))))))))...- ( -41.70, z-score = -2.05, R) >consensus UCAUCAUCGUCAUCGAUGAUGGCCAACCGACGCCGCUUGGCCGGCUCGCC_________AUCGACUUUGGCUGGCUUUUUGCGAGCCAAAAAGUUGGCCUCCUCGUCGCUGUCCUCC ..(((((((....))))))).(((((.((....)).)))))((((.................((((((((((.((.....)).))))))..))))(((......)))))))...... (-32.18 = -30.97 + -1.21)
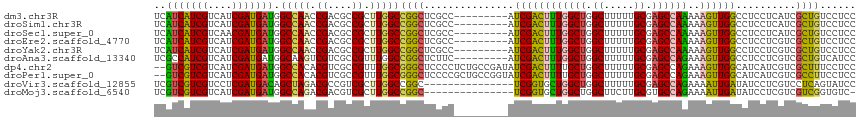
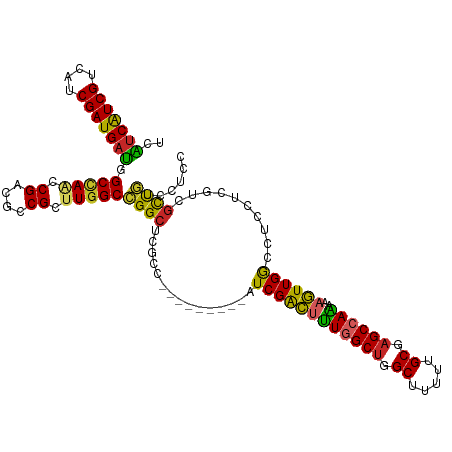
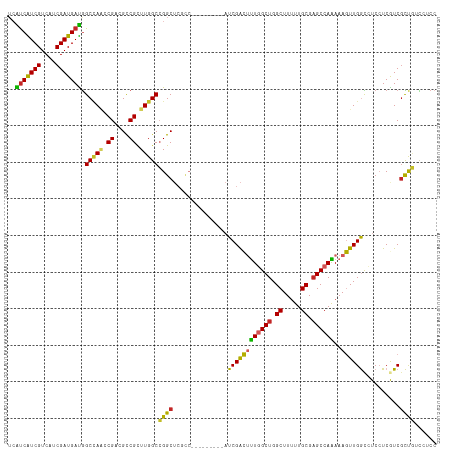
| Location | 8,989,410 – 8,989,518 |
|---|---|
| Length | 108 |
| Sequences | 10 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.69 |
| Shannon entropy | 0.36361 |
| G+C content | 0.57984 |
| Mean single sequence MFE | -41.50 |
| Consensus MFE | -21.46 |
| Energy contribution | -22.02 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.830451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8989410 108 + 27905053 ACAGCGAUGAGGAGGCCAACUUUUUGGCUCGCAAAAAGCCAGCCAAAGUCGAU---------GGCGAGCCGGCCAAGCGGCGUCGGUUGGCCAUCAUCGAUGACGAUGAUGAGGAUG .((.((((((...((((((((.(((((((.((.....)).)))))))((((.(---------(((......))))..))))...)))))))).)))))).))............... ( -45.60, z-score = -3.05, R) >droSim1.chr3R 15079433 108 - 27517382 ACAGCGAUGAGGAGGCCAACUUUUUGGCUCGCAAAAAGCCAGCCAAAGUCGAU---------GGCGAGCCGGCCAAGCGGCGUCGGUUGGCCAUCAUCGAUGACGAUGAUGAGGAUG .((.((((((...((((((((.(((((((.((.....)).)))))))((((.(---------(((......))))..))))...)))))))).)))))).))............... ( -45.60, z-score = -3.05, R) >droSec1.super_0 8105537 108 - 21120651 ACAGCGAUGAGGAGGCCAACUUUUUGGCUCGCAAAAAGCCAGCCAAAGUCGAU---------GGCGAGCCGGCCAAGCGGCGUCGGUUGGCCAUCAUCGUUGACGAUGAUGAGGAUG .(((((((((...((((((((.(((((((.((.....)).)))))))((((.(---------(((......))))..))))...)))))))).)))))))))............... ( -49.80, z-score = -4.11, R) >droEre2.scaffold_4770 5085281 108 - 17746568 ACAGCGACGAGGAGGCCAACUUUUUGGCUCGCAAAAAGCCAGCCAAAGUCGAU---------GGCGAGCCGGCCAAGCGGCGUCGGUUGGCCAUCAUCGAUGACGAUGAUGAGGAUG ...(((((((((((.....)))))))..)))).....(((((((...((((.(---------(((......))))..))))...)))))))((((((((....))))))))...... ( -45.60, z-score = -2.94, R) >droYak2.chr3R 13284395 108 + 28832112 ACAGCGACGAGGAGGCCAACUUUUUGGCUCGCAAAAAGCCAGCCAAAGUCGAU---------GGCGAGCCGGCCAAGCGGCGUCGGUUGGCCAUCAUCGAUGACGAUGAUGAGGAUG ...(((((((((((.....)))))))..)))).....(((((((...((((.(---------(((......))))..))))...)))))))((((((((....))))))))...... ( -45.60, z-score = -2.94, R) >droAna3.scaffold_13340 1540205 108 + 23697760 ACAGCGACGAGGAGGCCAACUUUCUGGCUCGCAAAAAGCCAGCCAAAGUCGAU---------GAAGAGCCGGCCAAACGGCGACGACUUGCCAUCAUCGAUGACGAUGGCGAGGAUG .((.((((.((((((....))))))((((.((.....)).))))...)))).)---------)....((((......)))).....(((((((((.((...)).))))))))).... ( -41.80, z-score = -3.15, R) >dp4.chr2 19689717 114 - 30794189 AAAGCGACGAUGAUGCCAACUUUCUGGCUCGCAAAAAGCCAGCAAAAGUCGAUAUCGGCAGAGGGGAGCCCGCCAAACGGCGACGUGUGGCCAUCAUCGAUGACGACGACGAUG--- ...(((.(((((((((((.((((((.(((.((.....)).)))....((((....)))))))))).....((((....)))).....))).)))))))).)).)..........--- ( -38.30, z-score = -1.34, R) >droPer1.super_0 9365202 114 + 11822988 AAGGCGACGAUGAUGCCAACUUUCUGGCUCGCAAAAAGCCAGCAAAAGUCGAUACCGGCAGCGGGGAGCCCGCCAAACGGCGACGUGUGGCCAUCAUCGAUGACGACGACGAUG--- ..(.((.((((((((((((((((((((((.......)))))).))).((((...(((...(((((...)))))....))))))))).))).)))))))).)).)..........--- ( -41.20, z-score = -1.48, R) >droVir3.scaffold_12855 6375200 102 - 10161210 CUGAGGACGAGGAUAUCAAUUUUCUGGCUCGCAAAAAGCCAGCCAGCACCGA---------------GCCGGCCAAGCGACGGCGUCUAGCUGUCAUCGAGGACGACGACGAUGAUG ....((((...............((((((.((.....)).))))))......---------------((((.........))))))))....(((((((.(.....)..))))))). ( -31.10, z-score = -0.16, R) >droMoj3.scaffold_6540 26064248 102 - 34148556 CCGACGACGAGGAUAUCAAUUUUCUGGCACGCAAGAAGCCAGCCAGCACCGA---------------GCCGGCCAAGCGACGUCGUCUGGCCAUCAUCGAUGACGACGACGACGACG ........((((((....))))))((((.(....)..))))(((.((.....---------------)).)))...(((.(((((((.(..((((...)))).)))))))).)).). ( -30.40, z-score = -0.58, R) >consensus ACAGCGACGAGGAGGCCAACUUUCUGGCUCGCAAAAAGCCAGCCAAAGUCGAU_________GGCGAGCCGGCCAAGCGGCGUCGGUUGGCCAUCAUCGAUGACGAUGACGAGGAUG .......(((.((........((((((((.((.....)).))))))))......................(((((..((....))..))))).)).))).................. (-21.46 = -22.02 + 0.56)
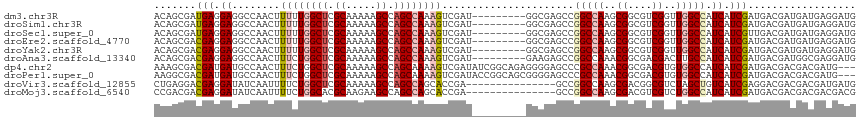
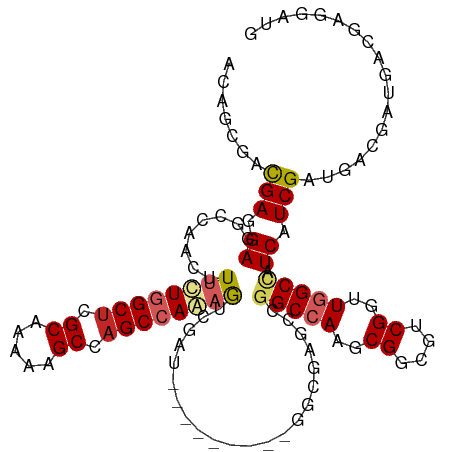
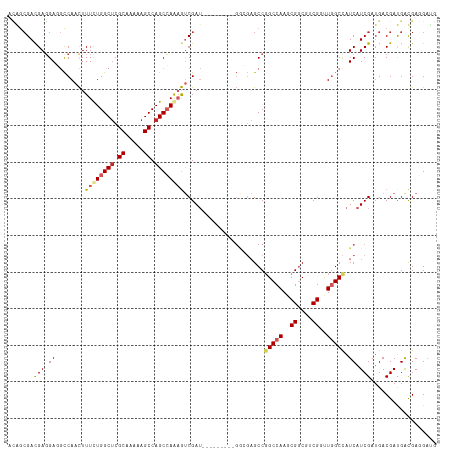
| Location | 8,989,410 – 8,989,518 |
|---|---|
| Length | 108 |
| Sequences | 10 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.69 |
| Shannon entropy | 0.36361 |
| G+C content | 0.57984 |
| Mean single sequence MFE | -42.97 |
| Consensus MFE | -34.31 |
| Energy contribution | -33.20 |
| Covariance contribution | -1.11 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.08 |
| SVM RNA-class probability | 0.997308 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8989410 108 - 27905053 CAUCCUCAUCAUCGUCAUCGAUGAUGGCCAACCGACGCCGCUUGGCCGGCUCGCC---------AUCGACUUUGGCUGGCUUUUUGCGAGCCAAAAAGUUGGCCUCCUCAUCGCUGU ...............((.((((((.((((((((((.((((......)))))))..---------......(((((((.((.....)).)))))))..)))))))...)))))).)). ( -43.10, z-score = -3.47, R) >droSim1.chr3R 15079433 108 + 27517382 CAUCCUCAUCAUCGUCAUCGAUGAUGGCCAACCGACGCCGCUUGGCCGGCUCGCC---------AUCGACUUUGGCUGGCUUUUUGCGAGCCAAAAAGUUGGCCUCCUCAUCGCUGU ...............((.((((((.((((((((((.((((......)))))))..---------......(((((((.((.....)).)))))))..)))))))...)))))).)). ( -43.10, z-score = -3.47, R) >droSec1.super_0 8105537 108 + 21120651 CAUCCUCAUCAUCGUCAACGAUGAUGGCCAACCGACGCCGCUUGGCCGGCUCGCC---------AUCGACUUUGGCUGGCUUUUUGCGAGCCAAAAAGUUGGCCUCCUCAUCGCUGU .......(((((((....)))))))((((((((((.((((......)))))))..---------......(((((((.((.....)).)))))))..)))))))............. ( -43.20, z-score = -3.56, R) >droEre2.scaffold_4770 5085281 108 + 17746568 CAUCCUCAUCAUCGUCAUCGAUGAUGGCCAACCGACGCCGCUUGGCCGGCUCGCC---------AUCGACUUUGGCUGGCUUUUUGCGAGCCAAAAAGUUGGCCUCCUCGUCGCUGU .....(((((((((....))))))))).....(((((......((((((((((..---------..))).(((((((.((.....)).)))))))..)))))))....))))).... ( -43.30, z-score = -3.22, R) >droYak2.chr3R 13284395 108 - 28832112 CAUCCUCAUCAUCGUCAUCGAUGAUGGCCAACCGACGCCGCUUGGCCGGCUCGCC---------AUCGACUUUGGCUGGCUUUUUGCGAGCCAAAAAGUUGGCCUCCUCGUCGCUGU .....(((((((((....))))))))).....(((((......((((((((((..---------..))).(((((((.((.....)).)))))))..)))))))....))))).... ( -43.30, z-score = -3.22, R) >droAna3.scaffold_13340 1540205 108 - 23697760 CAUCCUCGCCAUCGUCAUCGAUGAUGGCAAGUCGUCGCCGUUUGGCCGGCUCUUC---------AUCGACUUUGGCUGGCUUUUUGCGAGCCAGAAAGUUGGCCUCCUCGUCGCUGU .......(((((((((...))))))))).(((((..(((....))))))))...(---------(.((((...(((..((((((((.....))))))))..))).....)))).)). ( -43.40, z-score = -2.93, R) >dp4.chr2 19689717 114 + 30794189 ---CAUCGUCGUCGUCAUCGAUGAUGGCCACACGUCGCCGUUUGGCGGGCUCCCCUCUGCCGAUAUCGACUUUUGCUGGCUUUUUGCGAGCCAGAAAGUUGGCAUCAUCGUCGCUUU ---...((((((((....))))))))((((.(((....))).)))).(((........)))(((..((((((...(((((((.....))))))).))))))..)))........... ( -44.20, z-score = -2.68, R) >droPer1.super_0 9365202 114 - 11822988 ---CAUCGUCGUCGUCAUCGAUGAUGGCCACACGUCGCCGUUUGGCGGGCUCCCCGCUGCCGGUAUCGACUUUUGCUGGCUUUUUGCGAGCCAGAAAGUUGGCAUCAUCGUCGCCUU ---....(((((((....)))))))(((.((.((..((((...((((((...))))))..))))..)).....(((..((((((((.....))))))))..))).....)).))).. ( -45.90, z-score = -2.27, R) >droVir3.scaffold_12855 6375200 102 + 10161210 CAUCAUCGUCGUCGUCCUCGAUGACAGCUAGACGCCGUCGCUUGGCCGGC---------------UCGGUGCUGGCUGGCUUUUUGCGAGCCAGAAAAUUGAUAUCCUCGUCCUCAG .......(((((((....))))))).....(((((((((....)).))))---------------(((((.((((((.((.....)).))))))...))))).......)))..... ( -37.90, z-score = -1.93, R) >droMoj3.scaffold_6540 26064248 102 + 34148556 CGUCGUCGUCGUCGUCAUCGAUGAUGGCCAGACGACGUCGCUUGGCCGGC---------------UCGGUGCUGGCUGGCUUCUUGCGUGCCAGAAAAUUGAUAUCCUCGUCGUCGG ....((((((((((....))))))))))..(((((((......(((((((---------------.....)))))))(((.........)))................))))))).. ( -42.30, z-score = -1.88, R) >consensus CAUCCUCAUCAUCGUCAUCGAUGAUGGCCAACCGACGCCGCUUGGCCGGCUCGCC_________AUCGACUUUGGCUGGCUUUUUGCGAGCCAAAAAGUUGGCCUCCUCGUCGCUGU ......((((((((....))))))))(((((.((....)).)))))((((.................((((((((((.((.....)).))))))..))))(((......))))))). (-34.31 = -33.20 + -1.11)
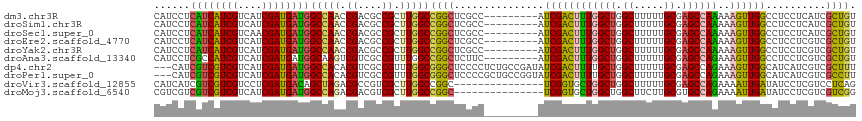
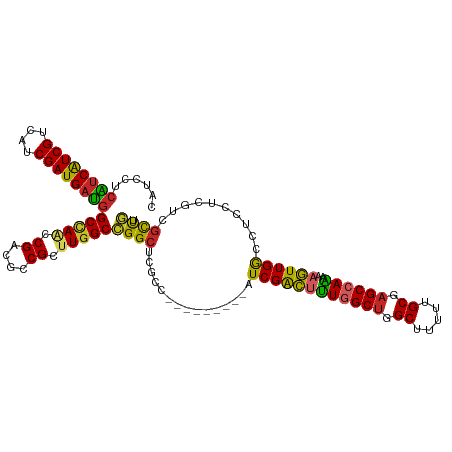
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:11:09 2011