
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,961,425 – 8,961,531 |
| Length | 106 |
| Max. P | 0.541927 |

| Location | 8,961,425 – 8,961,531 |
|---|---|
| Length | 106 |
| Sequences | 8 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 74.30 |
| Shannon entropy | 0.50447 |
| G+C content | 0.46564 |
| Mean single sequence MFE | -27.01 |
| Consensus MFE | -12.56 |
| Energy contribution | -12.31 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.541927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8961425 106 - 27905053 ACAAUUAUUUCCAGUGUAGCACCACACCAGCAUCGAGCGCAUGUCGCUCAC------AGAAGGCGAACUCUAUGAAGCCAAGGGUAAAGCGGAAGUUUAUUAAAACUACAGC ..............(((((......(((......(((((.....)))))..------....(((............)))...))).((((....)))).......))))).. ( -23.00, z-score = -0.47, R) >droSim1.chr3R 15059229 106 + 27517382 ACAAUUUUUUCCAGUGUAGCACCACACCAGCAUCGAGCGCAUGUCGCUCAC------AGAAGGCGAACUCUAUGAAGCCAAGGGUAAAGCGGAAGUUUAUUAAAACUACAGC ..............(((((......(((......(((((.....)))))..------....(((............)))...))).((((....)))).......))))).. ( -23.00, z-score = -0.39, R) >droSec1.super_0 8085455 106 + 21120651 ACAAUUUUUUCCAGUGAAGCACCACACCAGCAUCGAGCUCAUGUCGCUCAC------AGAAGGCGAACUCUAUGAAGCCAAGGGUAAAGCGGAAGUUUAUUAAAACUACAGC .........(((.((((.((...(((..(((.....)))..))).))))))------.....((..(((((.((....)))))))...)))))(((((....)))))..... ( -21.40, z-score = -0.15, R) >droYak2.chr3R 13264033 106 - 28832112 AGAAUUAUUUCCAGUGCAGCCCUACACCAGCAACGAGCGCAUGUCGCUCAC------AGAGGGCAAACUACGUGAAGCCAAGGAUAAAGCGGAAGUUUAUUAAAACUACAGC ..(((.(((((((((...(((((.(....)....(((((.....)))))..------..)))))..)))..((....(....).....))))))))..)))........... ( -21.30, z-score = 0.31, R) >droEre2.scaffold_4770 5064997 106 + 17746568 AUAAUUAUUUCCAGUGUAGCACCACACCAGCAUCGAGCGCAUGUCGCUCAC------AGAAGGCGAACUCCAUGAAGGGGAGGGUAAAGCGGAAGUUUAUUAAAACUACUGC .......(((((.(((......))).((..(((.((((((...((......------.))..)))..))).)))..))))))).....((((.(((((....))))).)))) ( -25.50, z-score = -0.50, R) >dp4.chr2 19669762 106 + 30794189 ------AUUGCCAGAGUGAGGAUUCGCCAUUAUCGAGUCCAUGUCGCUCACCCAAGGUGAAGGCGAACUCUAUAAAGCCAACGGCAAAGCGGAAGUUUAUUAAAAAGGCAGC ------.((((((((((..(((((((.......)))))))...(((((((((...)))))..))))))))).....(((...))).((((....))))........))))). ( -37.50, z-score = -3.93, R) >droPer1.super_0 9345316 106 - 11822988 ------AUUGCCAGAGUGAGGAUUCGCCAUUAUCGAGUCCAUGUCGCUCACCCAAGGUGAAGGCGAACUCUAUAAAGCCAACGGCAAAGCGGAAGUUUAUUAAAAAGGCAGC ------.((((((((((..(((((((.......)))))))...(((((((((...)))))..))))))))).....(((...))).((((....))))........))))). ( -37.50, z-score = -3.93, R) >droMoj3.scaffold_6540 26037493 95 + 34148556 ---------UCCAAUGAAG----UUGCUGGCAUUGAAUGCAUGUCAGAAGC---AACGGGCGGCAGACAGCAUAAAACUUUC-ACAAAGCGGAAGUUUAUUAAAACAGCAGC ---------.........(----((((((...((((((((.((((.(..((---.....))..).)))))))).((((((((-.......)))))))).))))..))))))) ( -26.90, z-score = -2.28, R) >consensus A_AAUUAUUUCCAGUGUAGCACCACACCAGCAUCGAGCGCAUGUCGCUCAC______AGAAGGCGAACUCUAUGAAGCCAAGGGUAAAGCGGAAGUUUAUUAAAACUACAGC .............(((......)))(((......((((.......))))............(((............)))...))).((((....)))).............. (-12.56 = -12.31 + -0.25)

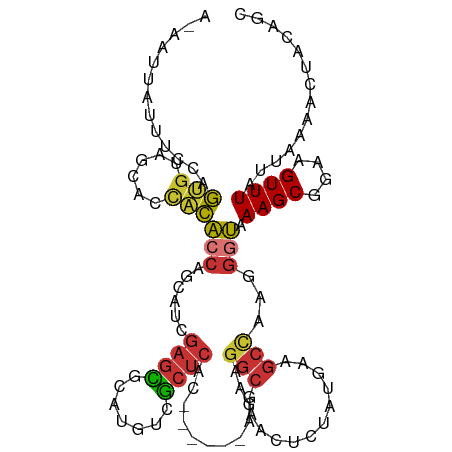
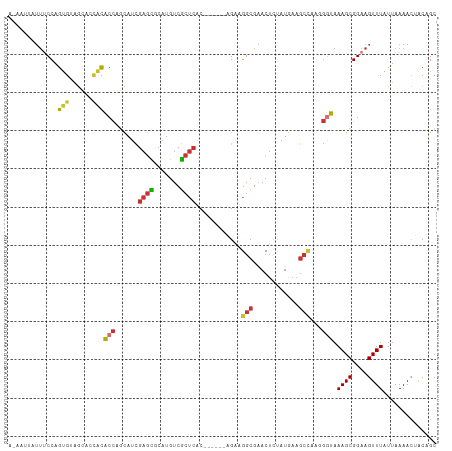
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:11:04 2011