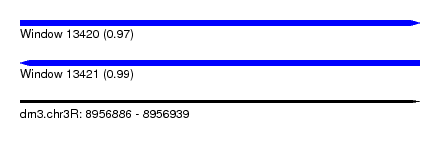
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,956,886 – 8,956,939 |
| Length | 53 |
| Max. P | 0.991807 |

| Location | 8,956,886 – 8,956,939 |
|---|---|
| Length | 53 |
| Sequences | 6 |
| Columns | 66 |
| Reading direction | forward |
| Mean pairwise identity | 62.54 |
| Shannon entropy | 0.69911 |
| G+C content | 0.42695 |
| Mean single sequence MFE | -14.67 |
| Consensus MFE | -9.31 |
| Energy contribution | -9.07 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.56 |
| Mean z-score | -0.82 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.972915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
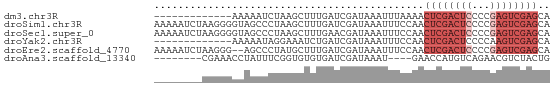
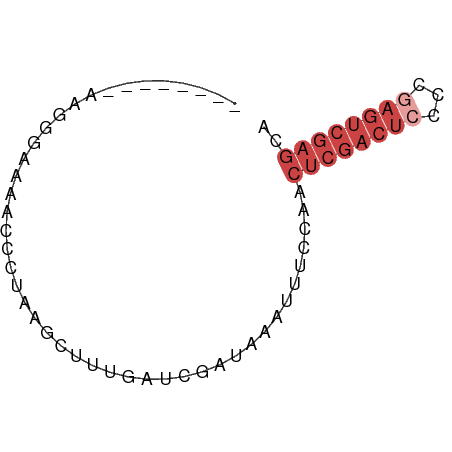
>dm3.chr3R 8956886 53 + 27905053 UGCUCGACUCGGGGAGUCGAGUUUUAAAUUUAUCGAUCAAAGCUUAGAUUUUU------------- .(((((((((...)))))))))............((((........))))...------------- ( -13.20, z-score = -1.62, R) >droSim1.chr3R 15054677 66 - 27517382 UGCUCGACUCGGGGAGUCGAGUUGGAAAUUUAUCGAUCAAAGCUUAGGGCUACCCCUUAGAUUUUU .(((((((((...)))))))))....................((.((((....)))).))...... ( -18.00, z-score = -0.72, R) >droSec1.super_0 8080909 66 - 21120651 UGCUCGACUCGGGGAGUCGAGUUGGAAAUUUAUCGUUCAAAGCUUAGGGCUACCCCUUAGAUUUUU .(((((((((...))))))))).(((((((((..((.....))..((((....))))))))))))) ( -18.50, z-score = -0.90, R) >droYak2.chr3R 13259513 53 + 28832112 UGCUCGACUUGGGGAGUCGAGUUGGAAAUUUAUCGAUCAGAUUUCCUAUUUUU------------- .(((((((((...))))))))).((((((((.......)))))))).......------------- ( -17.10, z-score = -2.72, R) >droEre2.scaffold_4770 5060433 64 - 17746568 UGCUCGACUCGGGGAGUCGAGUUGGAAAUUUAUCGAUCAAAGCAUAGGGCU--CCCUUAGAUUUUU .(((((((((...))))))))).(((((((((.............(((...--.)))))))))))) ( -16.71, z-score = -0.58, R) >droAna3.scaffold_13340 1510398 54 + 23697760 CAGUAGACGUUCUGACAUGGUUC----AUUUAUCGAUCACACACCGAAAUAGGUUUCG-------- (((........)))...(((((.----.......)))))...(((......)))....-------- ( -4.50, z-score = 1.65, R) >consensus UGCUCGACUCGGGGAGUCGAGUUGGAAAUUUAUCGAUCAAAGCUUAGAGCUACCCCUU________ .(((((((((...)))))))))............................................ ( -9.31 = -9.07 + -0.25)

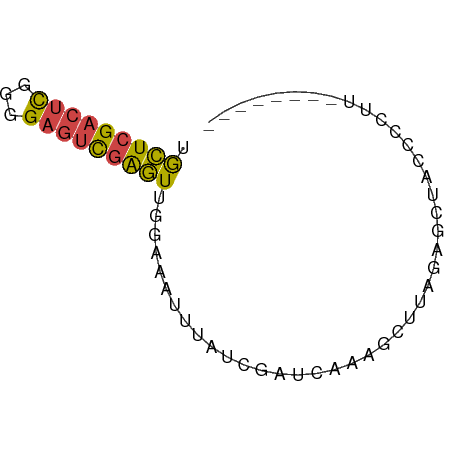
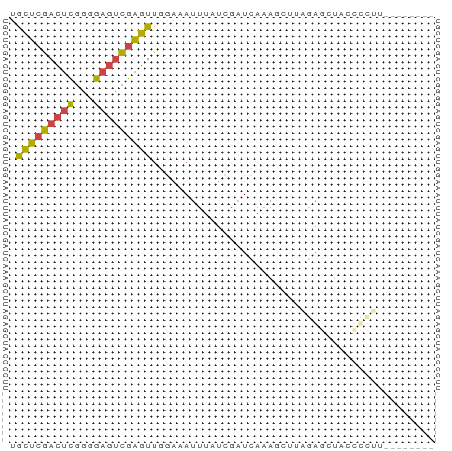
| Location | 8,956,886 – 8,956,939 |
|---|---|
| Length | 53 |
| Sequences | 6 |
| Columns | 66 |
| Reading direction | reverse |
| Mean pairwise identity | 62.54 |
| Shannon entropy | 0.69911 |
| G+C content | 0.42695 |
| Mean single sequence MFE | -13.25 |
| Consensus MFE | -6.23 |
| Energy contribution | -7.73 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.991807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8956886 53 - 27905053 -------------AAAAAUCUAAGCUUUGAUCGAUAAAUUUAAAACUCGACUCCCCGAGUCGAGCA -------------................................((((((((...)))))))).. ( -11.20, z-score = -2.73, R) >droSim1.chr3R 15054677 66 + 27517382 AAAAAUCUAAGGGGUAGCCCUAAGCUUUGAUCGAUAAAUUUCCAACUCGACUCCCCGAGUCGAGCA ..(((.((.((((....)))).)).))).................((((((((...)))))))).. ( -17.30, z-score = -1.92, R) >droSec1.super_0 8080909 66 + 21120651 AAAAAUCUAAGGGGUAGCCCUAAGCUUUGAACGAUAAAUUUCCAACUCGACUCCCCGAGUCGAGCA ..(((.((.((((....)))).)).))).................((((((((...)))))))).. ( -17.30, z-score = -2.04, R) >droYak2.chr3R 13259513 53 - 28832112 -------------AAAAAUAGGAAAUCUGAUCGAUAAAUUUCCAACUCGACUCCCCAAGUCGAGCA -------------.......((((((...........))))))..(((((((.....))))))).. ( -13.20, z-score = -3.46, R) >droEre2.scaffold_4770 5060433 64 + 17746568 AAAAAUCUAAGGG--AGCCCUAUGCUUUGAUCGAUAAAUUUCCAACUCGACUCCCCGAGUCGAGCA ....(((....((--(((.....)))))....)))..........((((((((...)))))))).. ( -14.70, z-score = -1.58, R) >droAna3.scaffold_13340 1510398 54 - 23697760 --------CGAAACCUAUUUCGGUGUGUGAUCGAUAAAU----GAACCAUGUCAGAACGUCUACUG --------(((((....)))))(((..((.((((((...----......)))).)).))..))).. ( -5.80, z-score = 1.14, R) >consensus ________AAGGGAAAACCCUAAGCUUUGAUCGAUAAAUUUCCAACUCGACUCCCCGAGUCGAGCA .............................................((((((((...)))))))).. ( -6.23 = -7.73 + 1.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:11:02 2011