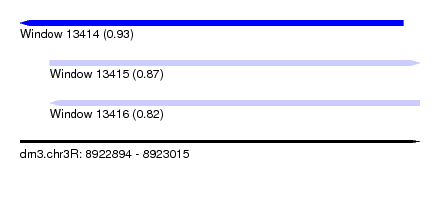
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,922,894 – 8,923,015 |
| Length | 121 |
| Max. P | 0.926547 |

| Location | 8,922,894 – 8,923,010 |
|---|---|
| Length | 116 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.61 |
| Shannon entropy | 0.11786 |
| G+C content | 0.34736 |
| Mean single sequence MFE | -21.51 |
| Consensus MFE | -19.59 |
| Energy contribution | -19.84 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.926547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8922894 116 - 27905053 GAAAACUUUGCGCCAACGGAAUGACAACAUUUUUGC-AAAGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUAAAGAAAUUUCCCACCACACAGCCAG--- ........((.((.....(((((((((((.....((-...)).....))))))))))).((((....(((((((.............)))))))....))))..........)))).--- ( -20.72, z-score = -1.93, R) >droSim1.chr3R 15017969 116 + 27517382 GAAAACUUUGCGCCAACGGAAUGACAACAUUUUUGC-AAAGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUAAAGAAAUUUCCCACCACACAGCCAG--- ........((.((.....(((((((((((.....((-...)).....))))))))))).((((....(((((((.............)))))))....))))..........)))).--- ( -20.72, z-score = -1.93, R) >droSec1.super_0 8046604 116 + 21120651 GAAAACUUUGCGCCAACGGAAUGACAACAUUUUUGC-AAAGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUAAAGAAAUUUCCCACCACACAGCCAG--- ........((.((.....(((((((((((.....((-...)).....))))))))))).((((....(((((((.............)))))))....))))..........)))).--- ( -20.72, z-score = -1.93, R) >droYak2.chr3R 13225261 116 - 28832112 GAAAACUUUGCGCCAACGGAAUGACAACAUUUUUGC-AAAGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUAAAGAAAUUUCCCACCACACUGCCAG--- ........((.((.....(((((((((((.....((-...)).....))))))))))).((((....(((((((.............)))))))....))))..........)))).--- ( -20.52, z-score = -1.68, R) >droEre2.scaffold_4770 5026953 116 + 17746568 GAAAACUUUGCGCCAACGGAAUGACAACAUUUUUGC-AAAGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUAAAGAAAUUUCCCACCACACAGCCAG--- ........((.((.....(((((((((((.....((-...)).....))))))))))).((((....(((((((.............)))))))....))))..........)))).--- ( -20.72, z-score = -1.93, R) >droAna3.scaffold_13340 1476555 116 - 23697760 CAAAAGUUUGCGCCAACGGAAUGACAACAUUUUUGC-AAAGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUAAAGAAAUUUCCCACCACACAGCCAG--- ........((.((.....(((((((((((.....((-...)).....))))))))))).((((....(((((((.............)))))))....))))..........)))).--- ( -20.72, z-score = -1.70, R) >dp4.chr2 19629252 119 + 30794189 AAAAACUUUGCGCCAACGGAAUGACAACAUUUUUGC-AAAGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUAAAGAAAUUUCCCACCACACAGCGGACAG .....((.(.(((.....(((((((((((.....((-...)).....))))))))))).((((....(((((((.............)))))))....))))..........))).).)) ( -22.92, z-score = -2.07, R) >droPer1.super_0 9305580 119 - 11822988 AAAAACUUUGCGCCAACGGAAUGACAACAUUUUUGC-AAAGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUAAAGAAAUUUCCCACCACACAGCGGACGG ..........((((....(((((((((((.....((-...)).....))))))))))).((((....(((((((.............)))))))....))))............)).)). ( -23.02, z-score = -1.68, R) >droWil1.scaffold_181108 1184137 116 + 4707319 CAAAACUUUACUCCAACGGAAUGACAACAUUUUUGC-AAAGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUAAAGAAAUUUCCCACCACACAGACCA--- ..................(((((((((((.....((-...)).....))))))))))).((((....(((((((.............)))))))....))))...............--- ( -18.82, z-score = -2.19, R) >droVir3.scaffold_12855 6303332 117 + 10161210 GAAAACUUUGCAGCAACGGAAUGACAACAUUUUUGC-AAAGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUAAAGAAAUUUCCCACCACAGAGCACAA-- .....(((((........(((((((((((.....((-...)).....))))))))))).((((....(((((((.............)))))))....))))......))))).....-- ( -22.12, z-score = -1.90, R) >droGri2.scaffold_14906 7689105 118 + 14172833 GAAAACUUUACAGCAGCGGAAUGACAACAUUUUUGCAAAAGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUAAAGAAAUUUCCCACCACUCAGCACAA-- ...............((.(((((((((((.....((....)).....))))))))))).((((....(((((((.............)))))))....))))..........))....-- ( -23.02, z-score = -2.62, R) >droMoj3.scaffold_6540 25985150 117 + 34148556 GAAAAGUUUGCAGCAACGGAAUGACAACAUUUUUGC-AAAGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUUAAGAAAUUUCCCACCACACAGCGCAA-- .......((((.((....(((((((((((.....((-...)).....))))))))))).((((....(((((((((..........)))))))))...))))..........))))))-- ( -24.10, z-score = -2.10, R) >consensus GAAAACUUUGCGCCAACGGAAUGACAACAUUUUUGC_AAAGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUAAAGAAAUUUCCCACCACACAGCCAA___ .........((.......(((((((((((.....((....)).....))))))))))).((((....(((((((.............)))))))....))))..........))...... (-19.59 = -19.84 + 0.25)
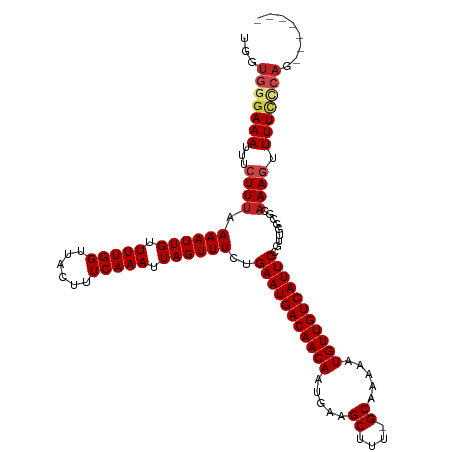
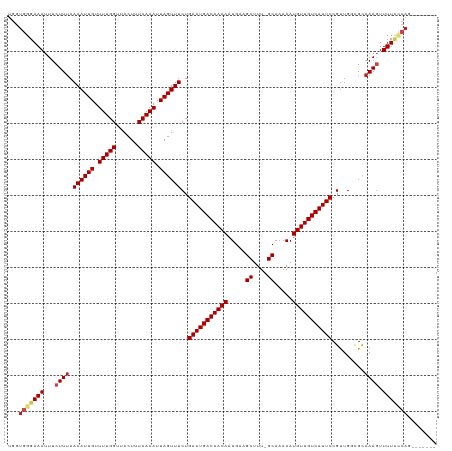
| Location | 8,922,903 – 8,923,015 |
|---|---|
| Length | 112 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 94.78 |
| Shannon entropy | 0.11560 |
| G+C content | 0.33327 |
| Mean single sequence MFE | -26.97 |
| Consensus MFE | -24.80 |
| Energy contribution | -25.51 |
| Covariance contribution | 0.71 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.874939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8922903 112 + 27905053 UGGUGGGAAAUUUCUUUAAAAUUGUUUUGGUUACUUUCAAAUUAGUUUCUGAAUGACAACAAUGAAGCUUU-GCAAAAAUGUUGUCAUUCCGUUGGCGCAAAGUUUUCCCAGA------ ...(((((((...((((.((((((.(((((......))))).))))))..(((((((((((.....((...-)).....))))))))))).........)))).)))))))..------ ( -29.00, z-score = -2.11, R) >droSim1.chr3R 15017978 112 - 27517382 UGGUGGGAAAUUUCUUUAAAAUUGUUUUGGUUACUUUCAAAUUAGUUUCUGAAUGACAACAAUGAAGCUUU-GCAAAAAUGUUGUCAUUCCGUUGGCGCAAAGUUUUCCCAGA------ ...(((((((...((((.((((((.(((((......))))).))))))..(((((((((((.....((...-)).....))))))))))).........)))).)))))))..------ ( -29.00, z-score = -2.11, R) >droSec1.super_0 8046613 112 - 21120651 UGGUGGGAAAUUUCUUUAAAAUUGUUUUGGUUACUUUCAAAUUAGUUUCUGAAUGACAACAAUGAAGCUUU-GCAAAAAUGUUGUCAUUCCGUUGGCGCAAAGUUUUCCCAGA------ ...(((((((...((((.((((((.(((((......))))).))))))..(((((((((((.....((...-)).....))))))))))).........)))).)))))))..------ ( -29.00, z-score = -2.11, R) >droYak2.chr3R 13225270 111 + 28832112 UGGUGGGAAAUUUCUUUAAAAUUGUUUUGGUUACUUUCAAAUUAGUUUCUGAAUGACAACAAUGAAGCUUU-GCAAAAAUGUUGUCAUUCCGUUGGCGCAAAGUUUUCCCAG------- ...(((((((...((((.((((((.(((((......))))).))))))..(((((((((((.....((...-)).....))))))))))).........)))).))))))).------- ( -29.00, z-score = -2.43, R) >droEre2.scaffold_4770 5026962 111 - 17746568 UGGUGGGAAAUUUCUUUAAAAUUGUUUUGGUUACUUUCAAAUUAGUUUCUGAAUGACAACAAUGAAGCUUU-GCAAAAAUGUUGUCAUUCCGUUGGCGCAAAGUUUUCCCAG------- ...(((((((...((((.((((((.(((((......))))).))))))..(((((((((((.....((...-)).....))))))))))).........)))).))))))).------- ( -29.00, z-score = -2.43, R) >droAna3.scaffold_13340 1476564 111 + 23697760 UGGUGGGAAAUUUCUUUAAAAUUGUUUUGGUUACUUUCAAAUUAGUUUCUGAAUGACAACAAUGAAGCUUU-GCAAAAAUGUUGUCAUUCCGUUGGCGCAAACUUUUGCCUU------- .((..(((..........((((((.(((((......))))).))))))..(((((((((((.....((...-)).....))))))))))).(((......))))))..))..------- ( -24.00, z-score = -0.91, R) >dp4.chr2 19629264 111 - 30794189 UGGUGGGAAAUUUCUUUAAAAUUGUUUUGGUUACUUUCAAAUUAGUUUCUGAAUGACAACAAUGAAGCUUU-GCAAAAAUGUUGUCAUUCCGUUGGCGCAAAGUUUUUGGAG------- .............(((((((((((((((((......)))))...(((.(.(((((((((((.....((...-)).....))))))))))).)..)))))))...))))))))------- ( -21.10, z-score = 0.18, R) >droPer1.super_0 9305592 111 + 11822988 UGGUGGGAAAUUUCUUUAAAAUUGUUUUGGUUACUUUCAAAUUAGUUUCUGAAUGACAACAAUGAAGCUUU-GCAAAAAUGUUGUCAUUCCGUUGGCGCAAAGUUUUUGGAG------- .............(((((((((((((((((......)))))...(((.(.(((((((((((.....((...-)).....))))))))))).)..)))))))...))))))))------- ( -21.10, z-score = 0.18, R) >droWil1.scaffold_181108 1184146 111 - 4707319 UGGUGGGAAAUUUCUUUAAAAUUGUUUUGGUUACUUUCAAAUUAGUUUCUGAAUGACAACAAUGAAGCUUU-GCAAAAAUGUUGUCAUUCCGUUGGAGUAAAGUUUUGCCAU------- (((..(((.....(((((((((((.(((((......))))).)))))...(((((((((((.....((...-)).....)))))))))))..)))))).....)))..))).------- ( -28.40, z-score = -2.52, R) >droVir3.scaffold_12855 6303342 118 - 10161210 UGGUGGGAAAUUUCUUUAAAAUUGUUUUGGUUACUUUCAAAUUAGUUUCUGAAUGACAACAAUGAAGCUUU-GCAAAAAUGUUGUCAUUCCGUUGCUGCAAAGUUUUCACAACCGGGCA .(((.((((((((............(((((......))))).((((..(.(((((((((((.....((...-)).....))))))))))).)..))))..))))))))...)))..... ( -28.20, z-score = -1.19, R) >droGri2.scaffold_14906 7689115 113 - 14172833 UGGUGGGAAAUUUCUUUAAAAUUGUUUUGGUUACUUUCAAAUUAGUUUCUGAAUGACAACAAUGAAGCUUUUGCAAAAAUGUUGUCAUUCCGCUGCUGUAAAGUUUUCUCAGU------ ...(((((((...(((((.......(((((......))))).((((..(.(((((((((((.....((....)).....))))))))))).)..))))))))).)))))))..------ ( -30.30, z-score = -3.23, R) >droMoj3.scaffold_6540 25985160 111 - 34148556 UGGUGGGAAAUUUCUUAAAAAUUGUUUUGGUUACUUUCAAAUUAGUUUCUGAAUGACAACAAUGAAGCUUU-GCAAAAAUGUUGUCAUUCCGUUGCUGCAAACUUUUCUCAA------- ...(((((((...............(((((......))))).((((..(.(((((((((((.....((...-)).....))))))))))).)..))))......))))))).------- ( -25.50, z-score = -2.01, R) >consensus UGGUGGGAAAUUUCUUUAAAAUUGUUUUGGUUACUUUCAAAUUAGUUUCUGAAUGACAACAAUGAAGCUUU_GCAAAAAUGUUGUCAUUCCGUUGGCGCAAAGUUUUCCCAG_______ ...(((((((...((((.((((((.(((((......))))).))))))..(((((((((((.....((....)).....))))))))))).........)))).)))))))........ (-24.80 = -25.51 + 0.71)

| Location | 8,922,903 – 8,923,015 |
|---|---|
| Length | 112 |
| Sequences | 12 |
| Columns | 119 |
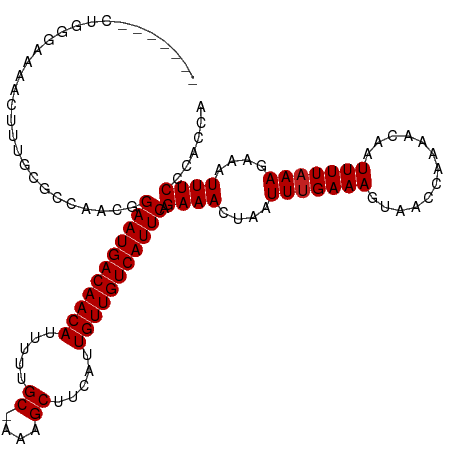
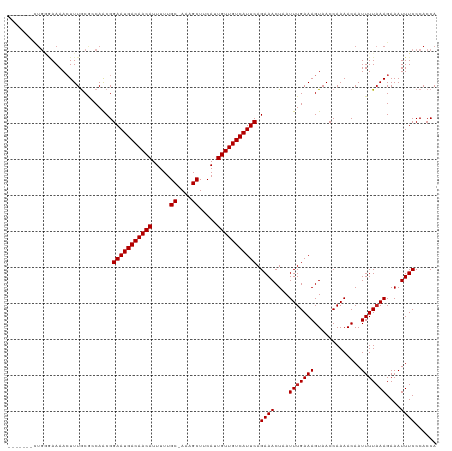
| Reading direction | reverse |
| Mean pairwise identity | 94.78 |
| Shannon entropy | 0.11560 |
| G+C content | 0.33327 |
| Mean single sequence MFE | -23.30 |
| Consensus MFE | -18.80 |
| Energy contribution | -18.89 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.822891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8922903 112 - 27905053 ------UCUGGGAAAACUUUGCGCCAACGGAAUGACAACAUUUUUGC-AAAGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUAAAGAAAUUUCCCACCA ------..(((((((.(((((........(((((((((((.....((-...)).....))))))))))).........((((........)))).......)))))...)))))))... ( -26.50, z-score = -2.93, R) >droSim1.chr3R 15017978 112 + 27517382 ------UCUGGGAAAACUUUGCGCCAACGGAAUGACAACAUUUUUGC-AAAGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUAAAGAAAUUUCCCACCA ------..(((((((.(((((........(((((((((((.....((-...)).....))))))))))).........((((........)))).......)))))...)))))))... ( -26.50, z-score = -2.93, R) >droSec1.super_0 8046613 112 + 21120651 ------UCUGGGAAAACUUUGCGCCAACGGAAUGACAACAUUUUUGC-AAAGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUAAAGAAAUUUCCCACCA ------..(((((((.(((((........(((((((((((.....((-...)).....))))))))))).........((((........)))).......)))))...)))))))... ( -26.50, z-score = -2.93, R) >droYak2.chr3R 13225270 111 - 28832112 -------CUGGGAAAACUUUGCGCCAACGGAAUGACAACAUUUUUGC-AAAGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUAAAGAAAUUUCCCACCA -------.(((((((.(((((........(((((((((((.....((-...)).....))))))))))).........((((........)))).......)))))...)))))))... ( -26.50, z-score = -2.97, R) >droEre2.scaffold_4770 5026962 111 + 17746568 -------CUGGGAAAACUUUGCGCCAACGGAAUGACAACAUUUUUGC-AAAGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUAAAGAAAUUUCCCACCA -------.(((((((.(((((........(((((((((((.....((-...)).....))))))))))).........((((........)))).......)))))...)))))))... ( -26.50, z-score = -2.97, R) >droAna3.scaffold_13340 1476564 111 - 23697760 -------AAGGCAAAAGUUUGCGCCAACGGAAUGACAACAUUUUUGC-AAAGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUAAAGAAAUUUCCCACCA -------..(((..........)))....(((((((((((.....((-...)).....))))))))))).((((....(((((((.............)))))))....))))...... ( -22.42, z-score = -1.76, R) >dp4.chr2 19629264 111 + 30794189 -------CUCCAAAAACUUUGCGCCAACGGAAUGACAACAUUUUUGC-AAAGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUAAAGAAAUUUCCCACCA -------......................(((((((((((.....((-...)).....))))))))))).((((....(((((((.............)))))))....))))...... ( -18.82, z-score = -1.69, R) >droPer1.super_0 9305592 111 - 11822988 -------CUCCAAAAACUUUGCGCCAACGGAAUGACAACAUUUUUGC-AAAGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUAAAGAAAUUUCCCACCA -------......................(((((((((((.....((-...)).....))))))))))).((((....(((((((.............)))))))....))))...... ( -18.82, z-score = -1.69, R) >droWil1.scaffold_181108 1184146 111 + 4707319 -------AUGGCAAAACUUUACUCCAACGGAAUGACAACAUUUUUGC-AAAGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUAAAGAAAUUUCCCACCA -------.(((..................(((((((((((.....((-...)).....))))))))))).((((....(((((((.............)))))))....)))))))... ( -20.62, z-score = -2.12, R) >droVir3.scaffold_12855 6303342 118 + 10161210 UGCCCGGUUGUGAAAACUUUGCAGCAACGGAAUGACAACAUUUUUGC-AAAGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUAAAGAAAUUUCCCACCA ......((((..(.....)..))))....(((((((((((.....((-...)).....))))))))))).((((....(((((((.............)))))))....))))...... ( -24.32, z-score = -1.31, R) >droGri2.scaffold_14906 7689115 113 + 14172833 ------ACUGAGAAAACUUUACAGCAGCGGAAUGACAACAUUUUUGCAAAAGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUAAAGAAAUUUCCCACCA ------..((.((((.(((((........(((((((((((.....((....)).....))))))))))).........((((........)))).......)))))...)))).))... ( -21.50, z-score = -1.99, R) >droMoj3.scaffold_6540 25985160 111 + 34148556 -------UUGAGAAAAGUUUGCAGCAACGGAAUGACAACAUUUUUGC-AAAGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUUAAGAAAUUUCCCACCA -------.((.(((((((((.(......)(((((((((((.....((-...)).....)))))))))))..)))))..(((((((((..........)))))))))...)))).))... ( -20.60, z-score = -1.19, R) >consensus _______CUGGGAAAACUUUGCGCCAACGGAAUGACAACAUUUUUGC_AAAGCUUCAUUGUUGUCAUUCAGAAACUAAUUUGAAAGUAACCAAAACAAUUUUAAAGAAAUUUCCCACCA .............................(((((((((((.....((....)).....))))))))))).((((....(((((((.............)))))))....))))...... (-18.80 = -18.89 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:10:58 2011