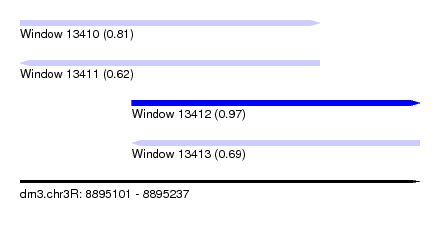
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,895,101 – 8,895,237 |
| Length | 136 |
| Max. P | 0.966838 |

| Location | 8,895,101 – 8,895,203 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 86.11 |
| Shannon entropy | 0.23564 |
| G+C content | 0.47509 |
| Mean single sequence MFE | -29.84 |
| Consensus MFE | -22.70 |
| Energy contribution | -22.50 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.810577 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8895101 102 + 27905053 AUCAAAAAGCAUAUUGAAAUUGGGUUAAUGUACGGAUACGGAGGCAACCAGUCCGGAUGCGAGUCCGUAUCCACAU------CCACAUCCGCAUCCAGUUGGUAUGGU .........(((((..(...(((((..((((..(((((((((.(((.((.....)).)))...)))))))))....------..))))....))))).)..))))).. ( -30.20, z-score = -1.48, R) >droEre2.scaffold_4770 4993353 79 - 17746568 AUCAAAAAGCAUAUUGAAAUUGGGUUAAUGUACGGAUACGGAGGCAACCAGUCCGGAUGCGAGUCCGUAUCCACAU------CCA----------------------- .((((........)))).....((...((((..(((((((((.(((.((.....)).)))...)))))))))))))------)).----------------------- ( -22.70, z-score = -1.83, R) >droYak2.chr3R 13191698 96 + 28832112 AUCAAAAAGCAUAUUGAAAUUGGGUUAAUGUACGGAUACGGAGGCAACCAGUCCGGAUGCGAGUCCGUAUCCACAU------CCACAUCC------AGUUGGCCUGGU .((((........)))).((..(((((((....((((..((((((.....))).(((((((....)))))))...)------))..))))------.)))))))..)) ( -29.70, z-score = -1.66, R) >droSec1.super_0 8019048 96 - 21120651 AUCAAAAAGCAUAUUGAAAUUGGGUUAAUGUACGGAUGCGGAGGCAACCAGUCCGGAUGCGAGUCCGUAUCCACAU------CCGCACCC------AGUUGGUAUGGU .........(((((..(...(((((..((((..(((((((((.(((.((.....)).)))...)))))))))))))------....))))------).)..))))).. ( -34.50, z-score = -2.75, R) >droSim1.chr3R 14989836 102 - 27517382 AUCAAAAAGCAUAUUGAAAUUGGGUUAAUGUACGGAAACGGAGGCAACCAGUCCGGAUGCGAGUCCGUAUCCACAUCCACAUCCGCACCC------AGUUGGUAUGGU .........(((((..(...(((((..((((..(....)((((((.....))).(((((((....)))))))...)))))))....))))------).)..))))).. ( -32.10, z-score = -2.00, R) >consensus AUCAAAAAGCAUAUUGAAAUUGGGUUAAUGUACGGAUACGGAGGCAACCAGUCCGGAUGCGAGUCCGUAUCCACAU______CCACACCC______AGUUGGUAUGGU .((((........))))...(((....((((..(((((((((.(((.((.....)).)))...)))))))))))))......)))....................... (-22.70 = -22.50 + -0.20)

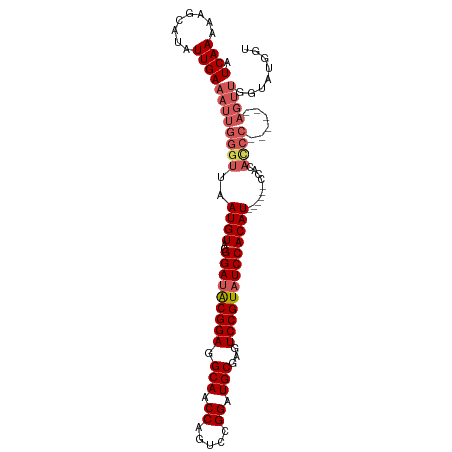
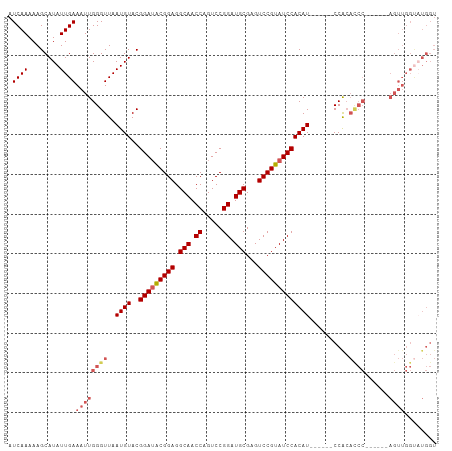
| Location | 8,895,101 – 8,895,203 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 86.11 |
| Shannon entropy | 0.23564 |
| G+C content | 0.47509 |
| Mean single sequence MFE | -28.36 |
| Consensus MFE | -21.50 |
| Energy contribution | -21.90 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.617230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8895101 102 - 27905053 ACCAUACCAACUGGAUGCGGAUGUGG------AUGUGGAUACGGACUCGCAUCCGGACUGGUUGCCUCCGUAUCCGUACAUUAACCCAAUUUCAAUAUGCUUUUUGAU ............(((((((((...((------((((((........))))))))((.(.....))))))))))))................((((........)))). ( -28.60, z-score = -1.10, R) >droEre2.scaffold_4770 4993353 79 + 17746568 -----------------------UGG------AUGUGGAUACGGACUCGCAUCCGGACUGGUUGCCUCCGUAUCCGUACAUUAACCCAAUUUCAAUAUGCUUUUUGAU -----------------------..(------(((((((((((((...(((.((.....)).))).)))))))))..))))).........((((........)))). ( -23.50, z-score = -2.21, R) >droYak2.chr3R 13191698 96 - 28832112 ACCAGGCCAACU------GGAUGUGG------AUGUGGAUACGGACUCGCAUCCGGACUGGUUGCCUCCGUAUCCGUACAUUAACCCAAUUUCAAUAUGCUUUUUGAU .((((.....))------))..((.(------(((((((((((((...(((.((.....)).))).)))))))))..))))).))......((((........)))). ( -30.10, z-score = -1.93, R) >droSec1.super_0 8019048 96 + 21120651 ACCAUACCAACU------GGGUGCGG------AUGUGGAUACGGACUCGCAUCCGGACUGGUUGCCUCCGCAUCCGUACAUUAACCCAAUUUCAAUAUGCUUUUUGAU .(((.......)------))((((((------(((((((..((((......))))....((...)))))))))))))))............((((........)))). ( -30.10, z-score = -2.16, R) >droSim1.chr3R 14989836 102 + 27517382 ACCAUACCAACU------GGGUGCGGAUGUGGAUGUGGAUACGGACUCGCAUCCGGACUGGUUGCCUCCGUUUCCGUACAUUAACCCAAUUUCAAUAUGCUUUUUGAU ...........(------((((...((((((....((((.(((((...(((.((.....)).))).))))).)))))))))).)))))...((((........)))). ( -29.50, z-score = -0.99, R) >consensus ACCAUACCAACU______GGAUGUGG______AUGUGGAUACGGACUCGCAUCCGGACUGGUUGCCUCCGUAUCCGUACAUUAACCCAAUUUCAAUAUGCUUUUUGAU ........................((......(((((((((((((...(((.((.....)).))).)))))))))..))))....))....((((........)))). (-21.50 = -21.90 + 0.40)

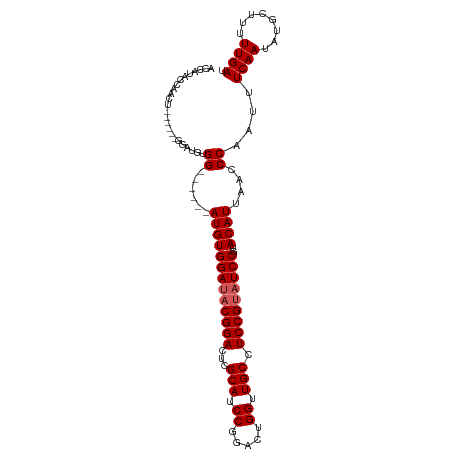
| Location | 8,895,139 – 8,895,237 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 91.70 |
| Shannon entropy | 0.11244 |
| G+C content | 0.57943 |
| Mean single sequence MFE | -39.43 |
| Consensus MFE | -34.70 |
| Energy contribution | -36.03 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.966838 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
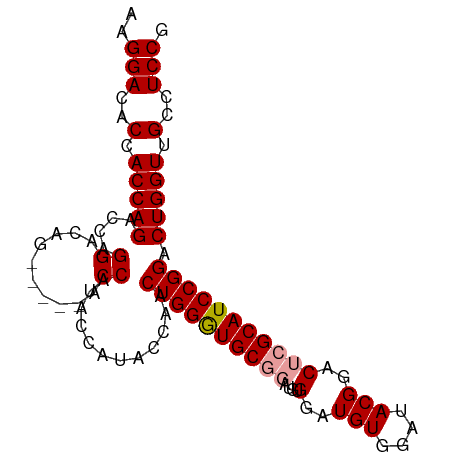
>dm3.chr3R 8895139 98 + 27905053 CGGAGGCAACCAGUCCGGAUGCGAGUCCGUAUCCACAUCCACAUCCGCAUCCAGUUGGUAUGGUAUGGUACGGUCUGUCCUGGUCUGGUGGUGUCCUU .(((.((.(((((.((((((...((.(((((.(((...(((.(((.((.....)).))).)))..)))))))).)))))).)).))))).)).))).. ( -41.60, z-score = -2.57, R) >droSec1.super_0 8019086 87 - 21120651 CGGAGGCAACCAGUCCGGAUGCGAGUCCGUAUCCACAUCC------GCACCCAGUUGGUAUGGUAUGGU-----CUGUCCUGGUCUGGUGGUGUCCUU .(((.((.(((((.(((((((((....))))))).(((((------(.(((.....))).))).)))..-----.......)).))))).)).))).. ( -37.70, z-score = -2.56, R) >droSim1.chr3R 14989874 93 - 27517382 CGGAGGCAACCAGUCCGGAUGCGAGUCCGUAUCCACAUCCACAUCCGCACCCAGUUGGUAUGGUAUGGU-----CUGUCCUGGUCUGGUGGUGUCCUU .(((.((.(((((.(((((((((....)))))))(((..(.((((((.(((.....))).))).)))).-----.)))...)).))))).)).))).. ( -39.00, z-score = -2.30, R) >consensus CGGAGGCAACCAGUCCGGAUGCGAGUCCGUAUCCACAUCCACAUCCGCACCCAGUUGGUAUGGUAUGGU_____CUGUCCUGGUCUGGUGGUGUCCUU .(((.((.(((((.(((((((((....))))))).((((((.(((.((.....)).))).))).)))..............)).))))).)).))).. (-34.70 = -36.03 + 1.33)


| Location | 8,895,139 – 8,895,237 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 91.70 |
| Shannon entropy | 0.11244 |
| G+C content | 0.57943 |
| Mean single sequence MFE | -33.87 |
| Consensus MFE | -27.52 |
| Energy contribution | -27.30 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.686667 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8895139 98 - 27905053 AAGGACACCACCAGACCAGGACAGACCGUACCAUACCAUACCAACUGGAUGCGGAUGUGGAUGUGGAUACGGACUCGCAUCCGGACUGGUUGCCUCCG ..(((..(.(((((.((.(((..(((((((((((((((((((..........)).))))).))))).)))))..))...))))).))))).)..))). ( -36.90, z-score = -2.62, R) >droSec1.super_0 8019086 87 + 21120651 AAGGACACCACCAGACCAGGACAG-----ACCAUACCAUACCAACUGGGUGC------GGAUGUGGAUACGGACUCGCAUCCGGACUGGUUGCCUCCG ..(((..(.(((((....((....-----.))............((((((((------(..(.((....)).)..))))))))).))))).)..))). ( -31.00, z-score = -1.87, R) >droSim1.chr3R 14989874 93 + 27517382 AAGGACACCACCAGACCAGGACAG-----ACCAUACCAUACCAACUGGGUGCGGAUGUGGAUGUGGAUACGGACUCGCAUCCGGACUGGUUGCCUCCG ..(((..(.(((((.((...(((.-----.(((((((.((((.....)))).)).))))).)))((((.((....)).)))))).))))).)..))). ( -33.70, z-score = -1.51, R) >consensus AAGGACACCACCAGACCAGGACAG_____ACCAUACCAUACCAACUGGGUGCGGAUGUGGAUGUGGAUACGGACUCGCAUCCGGACUGGUUGCCUCCG ..(((..(.(((((.((..............(((.(((.......)))))).......((((((((........)))))))))).))))).)..))). (-27.52 = -27.30 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:10:55 2011