
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,885,128 – 8,885,264 |
| Length | 136 |
| Max. P | 0.989160 |

| Location | 8,885,128 – 8,885,264 |
|---|---|
| Length | 136 |
| Sequences | 7 |
| Columns | 149 |
| Reading direction | forward |
| Mean pairwise identity | 75.54 |
| Shannon entropy | 0.46007 |
| G+C content | 0.39986 |
| Mean single sequence MFE | -33.73 |
| Consensus MFE | -17.70 |
| Energy contribution | -18.69 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.957554 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
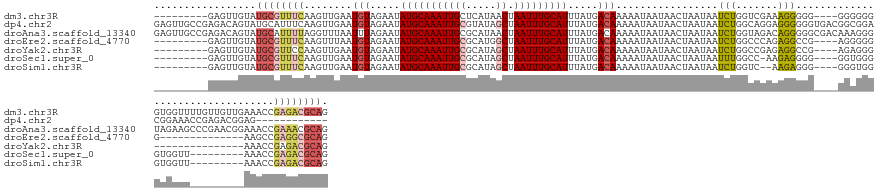
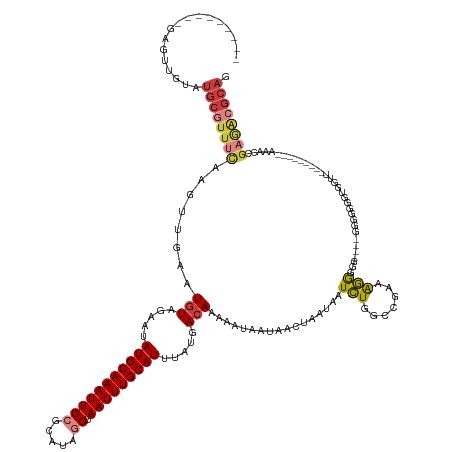
>dm3.chr3R 8885128 136 + 27905053 ---------GAGUUGUAUGCGUUUCAAGUUGAAUGUAGAAUAUGCAAAUUGCUCAUAACUAAUUUGCAUUUAUGACAAAAAUAAUAACUAAUAAUCUGGUCGAAAGGGGG----GGGGGGGUGGUUUUGUUGUUGAAACCGAGACGCAG ---------........((((((((........(((.....((((((((((........)))))))))).....))).........(((.....(((..((....))..)----))...)))((((((......)))))))))))))). ( -34.10, z-score = -2.23, R) >dp4.chr2 19588576 137 - 30794189 GAGUUGCCGAGACAGUAUGCAUUUCAAGUUGAAUGUAGAAUAUGCAAAUUGCGUAUAGCUAAUUUGCAUUUAUGACAAAAAUAAUAACUAAUAAUCUGGCAGGAGGGGGGUGACGGCGGACGGAAACCGAGACGGAG------------ ...(((((.(((.....(((((((......)))))))....(((((((((((.....)).))))))))).........................))))))))......(((..((.....))...))).........------------ ( -28.70, z-score = -0.36, R) >droAna3.scaffold_13340 1432243 149 + 23697760 GAGUUGCCGAGACAGUAUGCAUUUUAGGUUUAAUUUAGAAUAUGCAAAUUGCGCAUAACUAAUUUGCAUUUAUGACAAAAAUAAUAACUAAUAAUCUGGUAGACAGGGGGCGACAAAGGGUAGAAGCCCGAACGGAAACCGAAACGCAG ..((((((....((..(((((..((((((((......))))((((.......))))..))))..)))))...))....................((((.....)))).))))))...((((....))))...(((...)))........ ( -34.50, z-score = -1.97, R) >droEre2.scaffold_4770 4983338 122 - 17746568 ---------GAGUUGUAUGCGUUUCAAGUUUAAUGUAGAAUAUGCAAAUUGCGCAUGGCUAAUUUGCAUUUAUGACAAAAAUAAUAACUAAUAAUCUGGCCCAGAGGCCG----AGGGGGG--------------AAGCCGAGGCGCAG ---------........((((((((........(((.....(((((((((((.....)).))))))))).....))).................(((((((....)))).----))).((.--------------...)))))))))). ( -34.70, z-score = -1.58, R) >droYak2.chr3R 13181316 121 + 28832112 ---------GAGUUGUAUGCGUUCCAAGUUGAAUGUAGAAUAUGCAAAUUGCGCAUAGCUAAUUUGCAUUUAUGACAAAAAUAAUAACUAAUAAUCUGGCCGAGAGGCCG----AGAGGG---------------AAACCGAGACGCAG ---------........(((((..(........(((.....(((((((((((.....)).))))))))).....))).................(((((((....)))).----)))((.---------------...)))..))))). ( -39.30, z-score = -4.93, R) >droSec1.super_0 8009050 126 - 21120651 ---------GAGUUGUAUGCGUUUCAAGUUGAAUGUAGAAUAUGCAAAUUGCGCAUAGCUAAUUUGCAUUUAUGACAAAAAUAAUAACUAAUAAUUUGGCC-AAGAGGGG----GGUGGGGUGGUU---------AAACCGAGACGCAG ---------........((((((((........(((.....(((((((((((.....)).))))))))).....))).......((((((....(..(.((-.......)----).)..).)))))---------)....)))))))). ( -33.00, z-score = -2.23, R) >droSim1.chr3R 14979835 125 - 27517382 ---------GAGUUGUAUGCGUUUCAAGUUGAAUGUAGAAUAUGCAAAUUGCGCAUAGCUAAUUUGCAUUUAUGACAAAAAUAAUAACUAAUAAUCUGGUC--AAGAGGG----GGGUGGGUGGUU---------AAACCGAGACGCAG ---------........((((((((........(((.....(((((((((((.....)).))))))))).....))).........(((.....(((....--.)))...----.))).(((....---------..))))))))))). ( -31.80, z-score = -2.16, R) >consensus _________GAGUUGUAUGCGUUUCAAGUUGAAUGUAGAAUAUGCAAAUUGCGCAUAGCUAAUUUGCAUUUAUGACAAAAAUAAUAACUAAUAAUCUGGCCGAAAGGGGG____GGGGGGGUGGUU_________AAACCGAGACGCAG .................((((((((........(((.....(((((((((((.....)).))))))))).....))).................(((.......))).................................)))))))). (-17.70 = -18.69 + 0.98)
| Location | 8,885,128 – 8,885,264 |
|---|---|
| Length | 136 |
| Sequences | 7 |
| Columns | 149 |
| Reading direction | reverse |
| Mean pairwise identity | 75.54 |
| Shannon entropy | 0.46007 |
| G+C content | 0.39986 |
| Mean single sequence MFE | -25.17 |
| Consensus MFE | -15.44 |
| Energy contribution | -16.43 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.989160 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
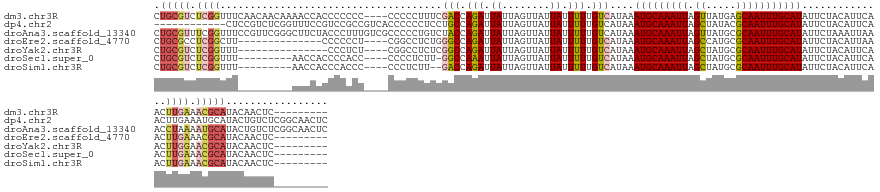
>dm3.chr3R 8885128 136 - 27905053 CUGCGUCUCGGUUUCAACAACAAAACCACCCCCCC----CCCCCUUUCGACCAGAUUAUUAGUUAUUAUUUUUGUCAUAAAUGCAAAUUAGUUAUGAGCAAUUUGCAUAUUCUACAUUCAACUUGAAACGCAUACAACUC--------- .(((((.(((((((........)))))........----.........(((.(((.((........)).))).)))....(((((((((.((.....)))))))))))................)).)))))........--------- ( -23.40, z-score = -3.24, R) >dp4.chr2 19588576 137 + 30794189 ------------CUCCGUCUCGGUUUCCGUCCGCCGUCACCCCCCUCCUGCCAGAUUAUUAGUUAUUAUUUUUGUCAUAAAUGCAAAUUAGCUAUACGCAAUUUGCAUAUUCUACAUUCAACUUGAAAUGCAUACUGUCUCGGCAACUC ------------....((..((((........))))..))........((((((((....(((...(((((((((.....(((((((((.((.....))))))))))).....)))........))))))...))))))).)))).... ( -25.20, z-score = -2.33, R) >droAna3.scaffold_13340 1432243 149 - 23697760 CUGCGUUUCGGUUUCCGUUCGGGCUUCUACCCUUUGUCGCCCCCUGUCUACCAGAUUAUUAGUUAUUAUUUUUGUCAUAAAUGCAAAUUAGUUAUGCGCAAUUUGCAUAUUCUAAAUUAAACCUAAAAUGCAUACUGUCUCGGCAACUC .(((((((.(((((......((((..(........)..)))).(((.....)))..........................(((((((((.((.....)))))))))))..........)))))..)))))))...(((....))).... ( -26.60, z-score = -0.71, R) >droEre2.scaffold_4770 4983338 122 + 17746568 CUGCGCCUCGGCUU--------------CCCCCCU----CGGCCUCUGGGCCAGAUUAUUAGUUAUUAUUUUUGUCAUAAAUGCAAAUUAGCCAUGCGCAAUUUGCAUAUUCUACAUUAAACUUGAAACGCAUACAACUC--------- .((((..((((((.--------------.....((----.((((....))))))......))))....((..(((.....(((((((((.((.....))))))))))).....)))..))....))..))))........--------- ( -26.00, z-score = -1.17, R) >droYak2.chr3R 13181316 121 - 28832112 CUGCGUCUCGGUUU---------------CCCUCU----CGGCCUCUCGGCCAGAUUAUUAGUUAUUAUUUUUGUCAUAAAUGCAAAUUAGCUAUGCGCAAUUUGCAUAUUCUACAUUCAACUUGGAACGCAUACAACUC--------- .(((((((.((((.---------------...(((----.((((....))))))).................(((.....(((((((((.((.....))))))))))).....)))...)))).)).)))))........--------- ( -29.30, z-score = -3.19, R) >droSec1.super_0 8009050 126 + 21120651 CUGCGUCUCGGUUU---------AACCACCCCACC----CCCCUCUU-GGCCAAAUUAUUAGUUAUUAUUUUUGUCAUAAAUGCAAAUUAGCUAUGCGCAAUUUGCAUAUUCUACAUUCAACUUGAAACGCAUACAACUC--------- .(((((.((((...---------..))........----.......(-(((.(((.((........)).))).))))...(((((((((.((.....)))))))))))................)).)))))........--------- ( -22.50, z-score = -2.34, R) >droSim1.chr3R 14979835 125 + 27517382 CUGCGUCUCGGUUU---------AACCACCCACCC----CCCUCUU--GACCAGAUUAUUAGUUAUUAUUUUUGUCAUAAAUGCAAAUUAGCUAUGCGCAAUUUGCAUAUUCUACAUUCAACUUGAAACGCAUACAACUC--------- .(((((.((((...---------..))........----......(--(((.(((.((........)).))).))))...(((((((((.((.....)))))))))))................)).)))))........--------- ( -23.20, z-score = -3.05, R) >consensus CUGCGUCUCGGUUU_________AACCACCCCCCC____CCCCCUUUCGGCCAGAUUAUUAGUUAUUAUUUUUGUCAUAAAUGCAAAUUAGCUAUGCGCAAUUUGCAUAUUCUACAUUCAACUUGAAACGCAUACAACUC_________ .(((((.((((.....................................(((.(((.((........)).))).)))....(((((((((.((.....)))))))))))..............)))).)))))................. (-15.44 = -16.43 + 0.98)
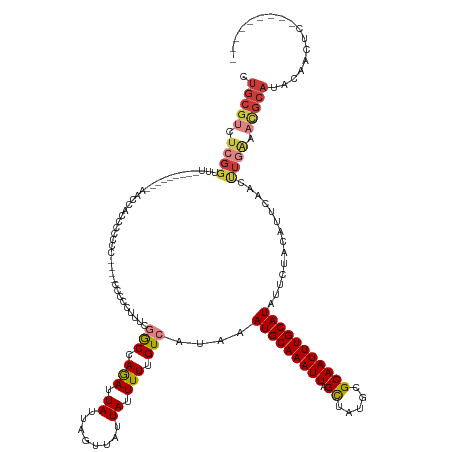
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:10:52 2011