| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,847,040 – 8,847,132 |
| Length | 92 |
| Max. P | 0.843990 |

| Location | 8,847,040 – 8,847,132 |
|---|---|
| Length | 92 |
| Sequences | 13 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 67.11 |
| Shannon entropy | 0.74571 |
| G+C content | 0.40239 |
| Mean single sequence MFE | -16.74 |
| Consensus MFE | -8.02 |
| Energy contribution | -8.35 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.15 |
| Mean z-score | -0.36 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.570657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8847040 92 + 27905053 AAGAGAUUGUUGUAAAACGAGACAUUUUACGCAAUCUGAGUUGGCG-CCGCACCCACGAUGAAUGUGAUCG-CAACUAAUGAAUGUUUUCUUUU---- ...(((((((.((((((.......))))))))))))).(((((.((-.((((..(.....)..))))..))-))))).................---- ( -18.00, z-score = 0.04, R) >droAna3.scaffold_13340 14364248 90 - 23697760 AAGAGAUUGUUGUAAAACAAGGCAUUUUACGCAAUCUGAGUUGGCG-CCGCACCAACGUCGGCUAUGUUCU-UUAUUAACCAAUUGUUAAUU------ ...(((((((.((((((.......))))))))))))).((((((((-.........)))))))).......-..((((((.....)))))).------ ( -19.30, z-score = -0.54, R) >droWil1.scaffold_181089 11583603 93 + 12369635 AAGAGAUUGUUGUAAAACGAGACAUUUUACGCAAUCUGAGUAGAAG-CCGCGCCUACACUGAACAAGAGACAUUACUAACCUAUU----ACCCCUUUA ...(((((((.((((((.......)))))))))))))..((((...-......))))............................----......... ( -12.30, z-score = -0.64, R) >droVir3.scaffold_12855 6192133 96 - 10161210 AAGAGAUUGUUGUAAAACGAAACAUUUUACGCAAUCUGAGACGACGACCCCGAC-ACGCUAGAUAUGUUUA-AUACUAAACAAAAUUAUUUUCCAAUU ...(((((((.((((((.......)))))))))))))((((...((....))..-..........((((((-....)))))).......))))..... ( -13.20, z-score = -1.31, R) >droGri2.scaffold_14906 7582597 93 - 14172833 AAGAGAUUGUUGUAAAACGAAACAUUUUACGCAAUCUGAGCAGACG-CAGCGACAGCGCUUGAUAUGUUUC-AUACUAAACGUUAUGUAUUCUUU--- ...(((((((.((((((.......)))))))))))))..(((((((-.((((....))))(((......))-).......)))).))).......--- ( -17.80, z-score = 0.17, R) >droMoj3.scaffold_6540 25872199 97 - 34148556 AAGAGAUUGUUGUAAAACGAAACAUUUUACGCAAUCUGAGACGAUGCCCGCAGUUGUGCUUGAUAUGUUUU-AUGCUAAUUGAAAUUCAUUUCGCAUU ...(((((((.((((((.......)))))))))))))((((.((......((((((.((.(((......))-).))))))))....)).))))..... ( -19.20, z-score = -0.42, R) >droPer1.super_0 9193191 95 + 11822988 AAGAGAUUGUUGUAAAACGAGACAUUUUACGCAAUCUGAGACGACG-ACGCGCCAAUGUCGAGCACGUUCC-CUACUAACCUAUUUGUAAAACCUUU- ...(((((((.((((((.......)))))))))))))(..(((.((-(((......)))))....)))..)-.........................- ( -15.90, z-score = -0.23, R) >dp4.chr2 19471416 95 - 30794189 AAGAGAUUGUUGUAAAACGAGACAUUUUACGCAAUCUGAGACGACG-ACGCGCCAAUGUCGAGCACGUUCC-CUACUAACCUAUUUGUAAAACCUUU- ...(((((((.((((((.......)))))))))))))(..(((.((-(((......)))))....)))..)-.........................- ( -15.90, z-score = -0.23, R) >droSim1.chr3R 14941723 92 - 27517382 AAGAGAUUGUUGUAAAACGAGACAUUUUACGCAAUCUGAGUUGGCG-CCGCACCCACGACGAAUGUGAUCG-CAACUAACCAAUGUUCUAUUUU---- ...(((((((.((((((.......))))))))))))).(((((.((-.((((..(.....)..))))..))-))))).................---- ( -17.70, z-score = -0.10, R) >droSec1.super_0 7971115 92 - 21120651 AAGAGAUUGUUGUAAAACGAGACAUUUUACGCAAUCUGAGUUGGCG-CCGCACCCACGACGAAUGUGAUCG-CAACUAACCAAUGUUCUAUUUU---- ...(((((((.((((((.......))))))))))))).(((((.((-.((((..(.....)..))))..))-))))).................---- ( -17.70, z-score = -0.10, R) >droYak2.chr3R 13142278 90 + 28832112 AAGAGAUUGUUGUAAAACGAGACAUUUUACGCAAUCUGAGUUGGCG-CCGCACCCACGUCGAAUGUGAUCG-CAACUAACCAGCGUUCUUUU------ ...(((((((.((((((.......)))))))))))))..(((((..-..((...(((((...)))))...)-)......)))))........------ ( -18.90, z-score = 0.23, R) >droEre2.scaffold_4770 4944803 90 - 17746568 AAGAGAUUGUUGUAAAACGAGACAUUUUACGCAAUCUGAGUUGGCG-CCGCACCCACGUCGAAUGUGAUCG-CAACUAACCAUCGUUCUUUU------ ...(((((((.((((((.......))))))))))))).(((((.((-.((((..(.....)..))))..))-)))))...............------ ( -17.70, z-score = 0.36, R) >apiMel3.GroupUn 7568196 72 + 399230636 --GGGUCAGUGCUCACAGUGUGAGUUUUAUCUAUCCAUAUCUAACCUUCUCUCACGUGAUUAAUGUUUUCCCUU------------------------ --(((.((.((.((((.(((.(((........................))).))))))).)).))....)))..------------------------ ( -14.06, z-score = -1.83, R) >consensus AAGAGAUUGUUGUAAAACGAGACAUUUUACGCAAUCUGAGUUGACG_CCGCACCCACGUCGAAUAUGUUCC_CUACUAACCAAUGUUCUAUUC_____ ...((((((.(((((((.......)))))))))))))............................................................. ( -8.02 = -8.35 + 0.33)

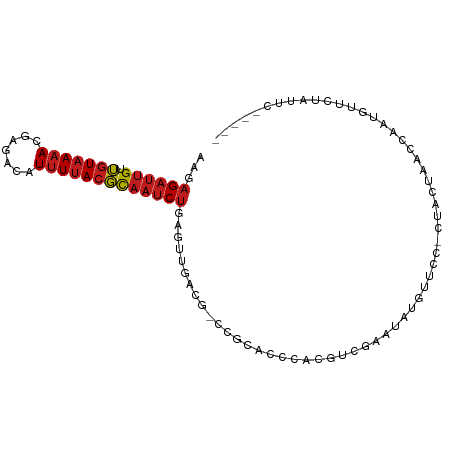
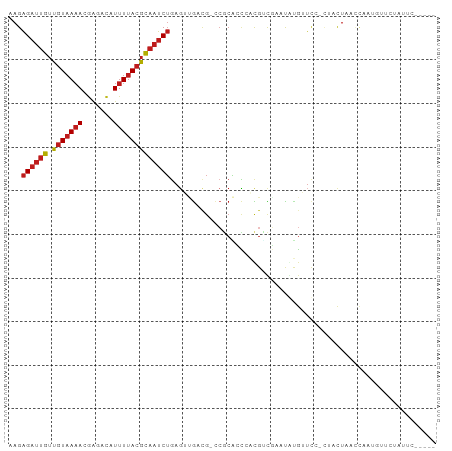
| Location | 8,847,040 – 8,847,132 |
|---|---|
| Length | 92 |
| Sequences | 13 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 67.11 |
| Shannon entropy | 0.74571 |
| G+C content | 0.40239 |
| Mean single sequence MFE | -20.20 |
| Consensus MFE | -8.40 |
| Energy contribution | -9.13 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.843990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8847040 92 - 27905053 ----AAAAGAAAACAUUCAUUAGUUG-CGAUCACAUUCAUCGUGGGUGCGG-CGCCAACUCAGAUUGCGUAAAAUGUCUCGUUUUACAACAAUCUCUU ----..((((...........(((((-((.((.(((((.....))))).))-)).)))))..(((((.((((((((...))))))))..))))))))) ( -21.30, z-score = -1.77, R) >droAna3.scaffold_13340 14364248 90 + 23697760 ------AAUUAACAAUUGGUUAAUAA-AGAACAUAGCCGACGUUGGUGCGG-CGCCAACUCAGAUUGCGUAAAAUGCCUUGUUUUACAACAAUCUCUU ------.........(((((((....-......))))))).(((((((...-)))))))..((((((.(((((((.....)))))))..))))))... ( -21.60, z-score = -1.68, R) >droWil1.scaffold_181089 11583603 93 - 12369635 UAAAGGGGU----AAUAGGUUAGUAAUGUCUCUUGUUCAGUGUAGGCGCGG-CUUCUACUCAGAUUGCGUAAAAUGUCUCGUUUUACAACAAUCUCUU ..(((((((----(((.(((.(((..((((((.........).)))))..)-))...)))...)))))((((((((...))))))))......))))) ( -19.10, z-score = -0.88, R) >droVir3.scaffold_12855 6192133 96 + 10161210 AAUUGGAAAAUAAUUUUGUUUAGUAU-UAAACAUAUCUAGCGU-GUCGGGGUCGUCGUCUCAGAUUGCGUAAAAUGUUUCGUUUUACAACAAUCUCUU .((((...........((((((....-))))))......(((.-(((.(((.......))).))))))((((((((...))))))))..))))..... ( -14.00, z-score = 0.05, R) >droGri2.scaffold_14906 7582597 93 + 14172833 ---AAAGAAUACAUAACGUUUAGUAU-GAAACAUAUCAAGCGCUGUCGCUG-CGUCUGCUCAGAUUGCGUAAAAUGUUUCGUUUUACAACAAUCUCUU ---..............((((((.((-(((((((.....((((.(((...(-(....))...))).))))...))))))))).))).)))........ ( -19.80, z-score = -1.07, R) >droMoj3.scaffold_6540 25872199 97 + 34148556 AAUGCGAAAUGAAUUUCAAUUAGCAU-AAAACAUAUCAAGCACAACUGCGGGCAUCGUCUCAGAUUGCGUAAAAUGUUUCGUUUUACAACAAUCUCUU .((((((((....)))).........-............(((....)))..))))......((((((.((((((((...))))))))..))))))... ( -16.00, z-score = -0.42, R) >droPer1.super_0 9193191 95 - 11822988 -AAAGGUUUUACAAAUAGGUUAGUAG-GGAACGUGCUCGACAUUGGCGCGU-CGUCGUCUCAGAUUGCGUAAAAUGUCUCGUUUUACAACAAUCUCUU -........................(-((((((((((.......)))))))-.....))))((((((.((((((((...))))))))..))))))... ( -20.50, z-score = -0.40, R) >dp4.chr2 19471416 95 + 30794189 -AAAGGUUUUACAAAUAGGUUAGUAG-GGAACGUGCUCGACAUUGGCGCGU-CGUCGUCUCAGAUUGCGUAAAAUGUCUCGUUUUACAACAAUCUCUU -........................(-((((((((((.......)))))))-.....))))((((((.((((((((...))))))))..))))))... ( -20.50, z-score = -0.40, R) >droSim1.chr3R 14941723 92 + 27517382 ----AAAAUAGAACAUUGGUUAGUUG-CGAUCACAUUCGUCGUGGGUGCGG-CGCCAACUCAGAUUGCGUAAAAUGUCUCGUUUUACAACAAUCUCUU ----...........(((((..((((-((..(((.......)))..)))))-))))))...((((((.((((((((...))))))))..))))))... ( -24.50, z-score = -1.85, R) >droSec1.super_0 7971115 92 + 21120651 ----AAAAUAGAACAUUGGUUAGUUG-CGAUCACAUUCGUCGUGGGUGCGG-CGCCAACUCAGAUUGCGUAAAAUGUCUCGUUUUACAACAAUCUCUU ----...........(((((..((((-((..(((.......)))..)))))-))))))...((((((.((((((((...))))))))..))))))... ( -24.50, z-score = -1.85, R) >droYak2.chr3R 13142278 90 - 28832112 ------AAAAGAACGCUGGUUAGUUG-CGAUCACAUUCGACGUGGGUGCGG-CGCCAACUCAGAUUGCGUAAAAUGUCUCGUUUUACAACAAUCUCUU ------..((((....((((..((((-((..(((.......)))..)))))-))))).....(((((.((((((((...))))))))..))))))))) ( -24.10, z-score = -0.99, R) >droEre2.scaffold_4770 4944803 90 + 17746568 ------AAAAGAACGAUGGUUAGUUG-CGAUCACAUUCGACGUGGGUGCGG-CGCCAACUCAGAUUGCGUAAAAUGUCUCGUUUUACAACAAUCUCUU ------..((((....((((..((((-((..(((.......)))..)))))-))))).....(((((.((((((((...))))))))..))))))))) ( -24.30, z-score = -1.16, R) >apiMel3.GroupUn 7568196 72 - 399230636 ------------------------AAGGGAAAACAUUAAUCACGUGAGAGAAGGUUAGAUAUGGAUAGAUAAAACUCACACUGUGAGCACUGACCC-- ------------------------...............((.(....).)).((((((.(((......)))...(((((...)))))..)))))).-- ( -12.40, z-score = -0.80, R) >consensus _____GAAUAAAACAUUGGUUAGUAG_CGAACAUAUUCGACGUGGGUGCGG_CGUCAACUCAGAUUGCGUAAAAUGUCUCGUUUUACAACAAUCUCUU ............................................((((....)))).....((((((.((((((((...))))))))..))))))... ( -8.40 = -9.13 + 0.73)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:10:50 2011