
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,841,427 – 8,841,536 |
| Length | 109 |
| Max. P | 0.933507 |

| Location | 8,841,427 – 8,841,536 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 73.33 |
| Shannon entropy | 0.50087 |
| G+C content | 0.50557 |
| Mean single sequence MFE | -31.47 |
| Consensus MFE | -17.47 |
| Energy contribution | -19.03 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.933507 |
| Prediction | RNA |
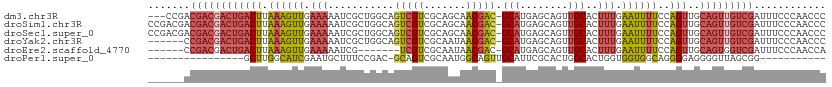
Download alignment: ClustalW | MAF
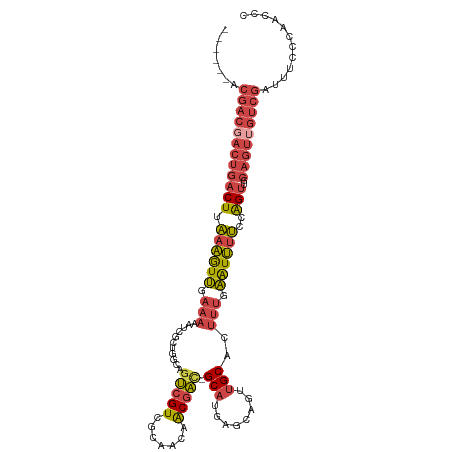
>dm3.chr3R 8841427 109 + 27905053 ---CCGACGACGACUGACUUAAAGUUGAAAAAUCGCUGGCAGUCGUCGCAGCAACGAC-GCAUGAGCAGUUGCACUUUGAAUUUUCCAGUUGCAGUUGUCGAUUUCCCAACCC ---.((((((((((((.((....(((....)))....))))))))))((((((((...-((....)).)))))...(((.......))).)))....))))............ ( -31.80, z-score = -1.65, R) >droSim1.chr3R 14936239 112 - 27517382 CCGACGACGACGACUGACUUAAAGUUGAAAAAUCGCUGGCAGUCGUCGCAGCAACGAC-GCAUGAGCAGUUGCACUUUGAAUUUUCCAGUUGCAGUUGUCGAUUUCCCAACCC .(((((((((((((((.((....(((....)))....))))))))))((((((((...-((....)).)))))...(((.......))).))).)))))))............ ( -36.90, z-score = -2.80, R) >droSec1.super_0 7965637 112 - 21120651 CCGACGACGACGACUGACUUAAAGUUGAAAAAUCGCUGGCAGUCGUCGCAGCAACGAC-GCAUGAGCAGUUGCACUUUGAAUUUUCCAGUUGCAGUUGUCGAUUUCCCAACCC .(((((((((((((((.((....(((....)))....))))))))))((((((((...-((....)).)))))...(((.......))).))).)))))))............ ( -36.90, z-score = -2.80, R) >droYak2.chr3R 13136653 106 + 28832112 ------CCGACGACUGACUUAAAGUUGAAAAAUCGCUGGCAGUCGUCGCAAUAACGAC-GCAUGAGCAGUUGCACUUUGAAUUUUCCAGUUGCAGUUGUCGAUUUCCCAACCC ------.(((((((((.....((.(.(((((.(((..(((((.(((((......))))-((....)).))))).)..))).))))).).)).)))))))))............ ( -29.10, z-score = -1.74, R) >droEre2.scaffold_4770 4939460 99 - 17746568 ------CCGACGACUGACUUAAAGUUGAAAAAUCG-------UCGUCGCAAUAACGAC-GCAUGAGCAGUUGCACUUUGAAUUUUCCAGUUGCAGUGGUCGAUUUCCCAACCA ------.((((.(((((((.((((((.(((..(((-------((((((......))))-).))))((....))..))).))))))..)))..)))).))))............ ( -26.80, z-score = -1.75, R) >droPer1.super_0 9188039 85 + 11822988 ----------------GCUUGGCAUCGAAUGCUUUCCGAC-GCAGUCGCAAUGGCAGUUGCAUUCGCACUGGCACUGGUGGUGGCAGGGGAGGGGUUAGCGG----------- ----------------.....((....(((.((((((...-((((..((....))..)))).......(((.(((.....))).))))))))).))).))..----------- ( -27.30, z-score = 0.55, R) >consensus ______ACGACGACUGACUUAAAGUUGAAAAAUCGCUGGCAGUCGUCGCAACAACGAC_GCAUGAGCAGUUGCACUUUGAAUUUUCCAGUUGCAGUUGUCGAUUUCCCAACCC .......((((((((((((.((((((.(((........((.(((((.......))))).))....((....))..))).))))))..)))..)))))))))............ (-17.47 = -19.03 + 1.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:10:48 2011