| Sequence ID | dm3.chr3R |
|---|---|
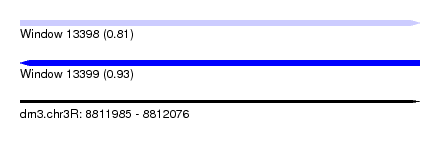
| Location | 8,811,985 – 8,812,076 |
| Length | 91 |
| Max. P | 0.926441 |

| Location | 8,811,985 – 8,812,076 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 72.37 |
| Shannon entropy | 0.56572 |
| G+C content | 0.68954 |
| Mean single sequence MFE | -44.91 |
| Consensus MFE | -26.71 |
| Energy contribution | -28.04 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.62 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.811720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
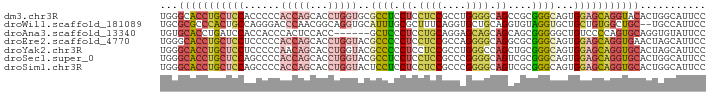
>dm3.chr3R 8811985 91 + 27905053 GGAAUGCCAGUGUACCUGCUCCACUGCCCGCGGCUGCCCCAGGCGGAGGAGGAGGCGCACCAGGUGCUGGUGGGGGUGGAGCAGGUGCCCA ((....)).(.((((((((((((((.((((((.((.((((.......)).)))).)))(((((...)))))))))))))))))))))).). ( -51.10, z-score = -1.78, R) >droWil1.scaffold_181089 11554190 89 + 12369635 GGAAUGGCA--GCAGCCACAGCAGCACCUACACCUGCAGCACCUGAAAGCGCAAAUGCACCUGCCGUUGGGUCCCUGGCAGUGGGCGCGCA ....((((.--...))))..((.((.(((((....(((((........))((....))...))).((..(....)..)).))))).)))). ( -28.80, z-score = 2.26, R) >droAna3.scaffold_13340 14335809 85 - 23697760 GGAAUACACCUGCACUGGGGACAGCCCCCGCUGCUGCUGCUCCUGCAGGAGGGAGC------GGUGGAGUGGGUGGUGGAUCAGGUGCACA ......((((((..(((....)))((((((((.(..((((((((......))))))------))..))))))).)).....)))))).... ( -44.90, z-score = -2.62, R) >droEre2.scaffold_4770 4909270 91 - 17746568 GGAAUGCUAGUUCACCUGCUCCACUGCCCGCGCCUGCCCCUGGCGGAGGAGGGGGCGUACCAGGUGCUGGUGGGGGAGGAGCAGGUGCCCA .........(..((((((((((.(..((.((((((((((((..(....).)))))).....)))))).))..)....))))))))))..). ( -48.40, z-score = -1.12, R) >droYak2.chr3R 13105888 91 + 28832112 GGAAUGCUAGUGCACCUGCUCCACUGCCCGCAGCUGGCCCAGGCGGAGGAGGGGGCGUACCAGGUGCUGUUGGGGGAGGAGCAGGUGCCCA .........(.(((((((((((....(((.((((..((((...(....)...))))((((...)))).)))).))).))))))))))).). ( -45.10, z-score = -0.54, R) >droSec1.super_0 7936351 91 - 21120651 GGAAUGCCAGUGCACCUGCUCCACUGCCCGCGACUGCCCCGGGCGGAGGAGGAGGCGUACCAGGUGCUGGUGGGGCUGGAGCAGGUGCCCA ((....)).(.(((((((((((.(((((((.(......))))))))......((.(.((((((...)))))).).))))))))))))).). ( -49.50, z-score = -1.62, R) >droSim1.chr3R 14907224 91 - 27517382 GGAAUGCCAGUGCACCUGCUCCACUGCCCGCGACUGCCCCGGGCGGAGGAGGAGGAGUACCAGGUGCUGGUGGGGCUGGAGCAGGUGCCCA ((....)).(.((((((((((((..((((((..((.((((.......)).))))..))(((((...))))).)))))))))))))))).). ( -46.60, z-score = -1.21, R) >consensus GGAAUGCCAGUGCACCUGCUCCACUGCCCGCGGCUGCCCCAGGCGGAGGAGGAGGCGUACCAGGUGCUGGUGGGGGUGGAGCAGGUGCCCA ...........(((((((((((...((((.((((.((.((..(((..........)))....)).)).)))))))).)))))))))))... (-26.71 = -28.04 + 1.34)



| Location | 8,811,985 – 8,812,076 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 72.37 |
| Shannon entropy | 0.56572 |
| G+C content | 0.68954 |
| Mean single sequence MFE | -41.36 |
| Consensus MFE | -25.25 |
| Energy contribution | -26.64 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.926441 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8811985 91 - 27905053 UGGGCACCUGCUCCACCCCCACCAGCACCUGGUGCGCCUCCUCCUCCGCCUGGGGCAGCCGCGGGCAGUGGAGCAGGUACACUGGCAUUCC .(((.(((((((((((.((((((((...)))))(((.((.((((.......)))).)).))))))..))))))))))).).))........ ( -43.50, z-score = -1.07, R) >droWil1.scaffold_181089 11554190 89 - 12369635 UGCGCGCCCACUGCCAGGGACCCAACGGCAGGUGCAUUUGCGCUUUCAGGUGCUGCAGGUGUAGGUGCUGCUGUGGCUGC--UGCCAUUCC .(((((((..(((((..(....)...))))).(((((((((((........)).)))))))))))))).)).(((((...--.)))))... ( -37.90, z-score = 0.42, R) >droAna3.scaffold_13340 14335809 85 + 23697760 UGUGCACCUGAUCCACCACCCACUCCACC------GCUCCCUCCUGCAGGAGCAGCAGCAGCGGGGGCUGUCCCCAGUGCAGGUGUAUUCC .(((((((((..............((.((------(((..((.((((....)))).)).)))))))((((....)))).)))))))))... ( -32.50, z-score = -0.98, R) >droEre2.scaffold_4770 4909270 91 + 17746568 UGGGCACCUGCUCCUCCCCCACCAGCACCUGGUACGCCCCCUCCUCCGCCAGGGGCAGGCGCGGGCAGUGGAGCAGGUGAACUAGCAUUCC .(..((((((((((.(.((((((((...))))).((((..(((((.....)))))..)))).)))..).))))))))))..)......... ( -45.20, z-score = -1.98, R) >droYak2.chr3R 13105888 91 - 28832112 UGGGCACCUGCUCCUCCCCCAACAGCACCUGGUACGCCCCCUCCUCCGCCUGGGCCAGCUGCGGGCAGUGGAGCAGGUGCACUAGCAUUCC ((((((((((((((.(..((..((((....((....))(((..........)))...))))..))..).))))))))))).)))....... ( -41.00, z-score = -1.31, R) >droSec1.super_0 7936351 91 + 21120651 UGGGCACCUGCUCCAGCCCCACCAGCACCUGGUACGCCUCCUCCUCCGCCCGGGGCAGUCGCGGGCAGUGGAGCAGGUGCACUGGCAUUCC .((((((((((((((((((.(((((...)))))..((...((.((((....)))).))..))))))..)))))))))))).))........ ( -45.60, z-score = -1.70, R) >droSim1.chr3R 14907224 91 + 27517382 UGGGCACCUGCUCCAGCCCCACCAGCACCUGGUACUCCUCCUCCUCCGCCCGGGGCAGUCGCGGGCAGUGGAGCAGGUGCACUGGCAUUCC .((((((((((((((((((((((((...))))).......((.((((....)))).))..).))))..)))))))))))).))........ ( -43.80, z-score = -1.53, R) >consensus UGGGCACCUGCUCCACCCCCACCAGCACCUGGUACGCCUCCUCCUCCGCCUGGGGCAGCCGCGGGCAGUGGAGCAGGUGCACUGGCAUUCC ...(((((((((((......(((((...)))))..((((.((.((((....)))).))....))))...)))))))))))........... (-25.25 = -26.64 + 1.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:10:43 2011