
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,780,843 – 8,780,974 |
| Length | 131 |
| Max. P | 0.819565 |

| Location | 8,780,843 – 8,780,974 |
|---|---|
| Length | 131 |
| Sequences | 12 |
| Columns | 154 |
| Reading direction | reverse |
| Mean pairwise identity | 73.81 |
| Shannon entropy | 0.54166 |
| G+C content | 0.44128 |
| Mean single sequence MFE | -32.00 |
| Consensus MFE | -16.13 |
| Energy contribution | -16.45 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.819565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8780843 131 - 27905053 ---CACC-CUUAACCCUUUGAUGCCCCCCGUGCU-GCAGUUGAUGUGACAGCG------------CCGCGCACUUUGUCAA-----UGGUG-UCUGUGAUAAAGUGUUUUAAAUUCCAAAUGCAUUCAACGUUAAAUGCGAAAAAGGUGAAAUG ---((((-.........(((((((.....(((((-(((.......)).)))))------------)...(((((((((((.-----.....-....)))))))))))..............))).))))(((.....))).....))))..... ( -31.30, z-score = -1.23, R) >droYak2.chr3R 13072883 132 - 28832112 ---CACCUCUUAACCCUUUGAUGCCCCCCGUGCU-GCAGUUGAUGUGACAGCG------------CCGCGCACUUUGUCAA-----UGGUG-UCUGUGAUAAAGUGUUUUAAAUUCCAAAUGCAUUCAACGUUAAAUGCGAAAAAGGUGAAAUA ---(((((.........(((((((.....(((((-(((.......)).)))))------------)...(((((((((((.-----.....-....)))))))))))..............))).))))(((.....)))....)))))..... ( -32.30, z-score = -1.41, R) >droEre2.scaffold_4770 4876492 131 + 17746568 ---CACCUCUUAACCCUUUGAUG-CCCCCGUGCU-GCAGUUGAUGUGACAGCG------------CCGCGCACUUUGUCAA-----UGGUG-UCUGUGAUAAAGUGUUUUAAAUUCCAAAUGCAUUCAACGUUAAAUGCGAAAAAGGUGAAAUA ---(((((.........((((((-(....(((((-(((.......)).)))))------------)...(((((((((((.-----.....-....)))))))))))..............))).))))(((.....)))....)))))..... ( -32.30, z-score = -1.32, R) >droSec1.super_0 7905594 131 + 21120651 ---CACC-CUUAACCCUUUGAUGCCCCCCGUGCU-GCAGUUGAUGUGACAGCG------------CCGCGCACUUUGUCAA-----UGGUG-UCUGUGAUAAAGUGUUUUAAAUUCCAAAUGCAUUCAACGUUAAAUGCGAAAAAGGUGAAAUG ---((((-.........(((((((.....(((((-(((.......)).)))))------------)...(((((((((((.-----.....-....)))))))))))..............))).))))(((.....))).....))))..... ( -31.30, z-score = -1.23, R) >droSim1.chr3R 14875558 131 + 27517382 ---CACC-CUUAA-CCUUUGAUGCCCCCCGUGGUGGCAGUUGAUGUGACAGCG------------CCGCGCACUUUGUCAA-----UGGUG-UCUGUGAUAAAGUGUUUUAAAUUCCAAAUGCAUUCAACGUUAAAUGCGAAAAGGGUGAAAUG ---((((-(((..-...(((((((........(((((.((((......)))))------------))))(((((((((((.-----.....-....)))))))))))..............))).))))(((.....)))..)))))))..... ( -40.30, z-score = -2.47, R) >droAna3.scaffold_13340 14304274 136 + 23697760 CACCCGCACACCGGCACACUCCUCUUCGUCCUGCUGCAGUUGAUGUGACAGCG------------CCGCGCACUUUGUC-AU----UGGUG-UCUGUGAUAAAGUGUUUUAAAUUCCAAAUGCAUUCAACGUUAAAUGCGAAAAAGGUGAAAUA ((((.(((...((((..((........))...((((((.......)).)))))------------))).((((((((((-((----.....-...)))))))))))).............)))......(((.....))).....))))..... ( -34.50, z-score = -0.30, R) >dp4.chr2 19392867 136 + 30794189 -GCCCCUCUGCCCCCUGCCUAUGCUCAUCCCUGUCGCAGUUGAUGUGACAGCG------------CCGCGCACUUUGUCAAU----GGGUG-UCUGUGAUAAAGUGUUUUAAAUUCCAAAUGCAUUCAACGCUAAAUGCAAAAAAGGUGAAAUA -.............................((((((((.....))))))))((------------((..(((((((((((.(----((...-.)))))))))))))).............((((((........)))))).....))))..... ( -34.10, z-score = -1.36, R) >droPer1.super_0 9116758 136 - 11822988 -CCGCCUGCCCCCCCUGACUAUGCUCAUCCCUGUCGCAGUUGAUGUGACAGCG------------CCGCGCACUUUGUCAAU----GGGUG-UCUGUGAUAAAGUGUUUUAAAUUCCAAAUGCAUUCAACGCUAAAUGCAAAAAAGGUGAAAUA -.(((((........(((..((((......((((((((.....))))))))..------------....(((((((((((.(----((...-.))))))))))))))..............)))))))..((.....)).....)))))..... ( -36.20, z-score = -1.97, R) >droWil1.scaffold_181089 11516481 118 - 12369635 ---------------------CUCCAAAAGUGACUGCAGUUGAUGUGACAGCG------------CCGCGCACUUUGUCAAU---CCUGUGCCCUCUGAUAAAGUGUUUUAAAUUCCAAAUGCAUUCAACGUUAAAUGCGAAAAAGGUGAAAUU ---------------------(.((.....(((.((((.(((..(((......------------.)))(((((((((((..---...........))))))))))).........))).)))).))).(((.....))).....)).)..... ( -25.12, z-score = -0.53, R) >droVir3.scaffold_12855 6122285 144 + 10161210 ---------UUUGUCCAGUUACGCAACCCCCUUCUGUAUUUGAUGUGUCAUCAGCGCUCCUUGCGCCGCGCACUUUGUCAACGUGCCUGUG-CCUGUGAUAAAGUGCUUUAAACUUAAAAUGCAUUCAACGUUAAAUGCAAAAAAGCUGAAUUG ---------......(((((..((................(((((...)))))((((.....)))).))(((((((((((.((.((....)-).))))))))))))).............((((((........))))))....)))))..... ( -38.20, z-score = -2.92, R) >droGri2.scaffold_14906 7511306 118 + 14172833 ---------ACUGCCCAGUAACG---CCCUUGUCUGCAUUUGAUGCAACAUCA--------------GCUCACUUUGUCAA------UGUG-CCUGUGAUAAAGUGAUUUAAACUCAAAACGCAUUCAACGUUAAAUGCAAAAAAGUCAAA--- ---------..(((....(((((---...(((..(((..((((((...)))))--------------).(((((((((((.------....-....)))))))))))..............)))..))))))))...)))...........--- ( -26.20, z-score = -1.83, R) >droMoj3.scaffold_6540 25794415 106 + 34148556 -----------------GUUACGC--CUCUUUUCUGCAUUUGAUGUGACAUCAG---------CGACGCGCACUUUGUCAA------UUCG-UCCGUGAUAAAGUGCUUUAAACUCGAAAUGCAUUCAACUUUAAACUCGA------------- -----------------.......--........(((((((((((...))))..---------......(((((((((((.------....-....)))))))))))..........))))))).................------------- ( -22.20, z-score = -0.64, R) >consensus ___CACC_CUUAACCCUUUGAUGCCCCCCGUGCUUGCAGUUGAUGUGACAGCG____________CCGCGCACUUUGUCAA_____UGGUG_UCUGUGAUAAAGUGUUUUAAAUUCCAAAUGCAUUCAACGUUAAAUGCGAAAAAGGUGAAAUA ...................................(((.(((((((.......................(((((((((((................))))))))))).............((....))))))))).)))............... (-16.13 = -16.45 + 0.32)
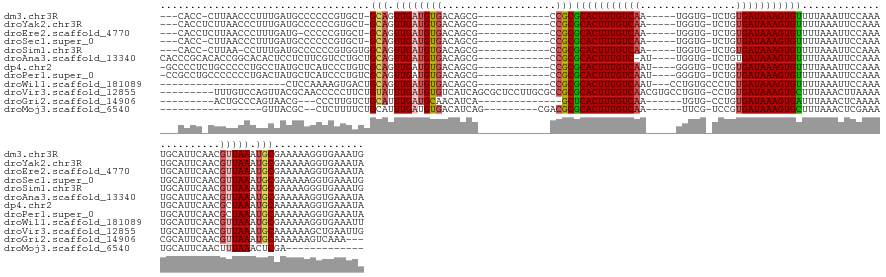
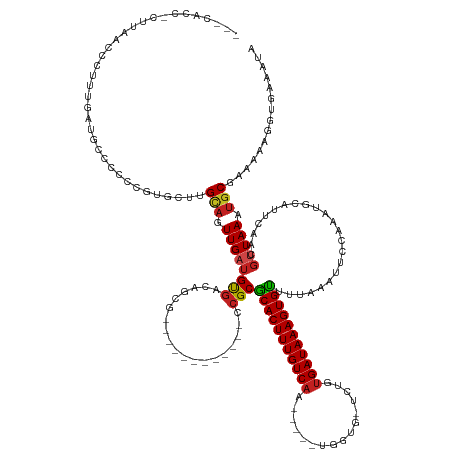

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:10:40 2011