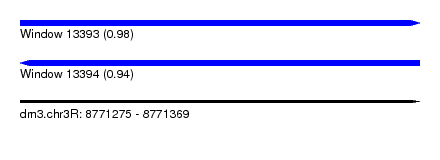
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,771,275 – 8,771,369 |
| Length | 94 |
| Max. P | 0.977446 |

| Location | 8,771,275 – 8,771,369 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 79.21 |
| Shannon entropy | 0.41111 |
| G+C content | 0.38542 |
| Mean single sequence MFE | -18.06 |
| Consensus MFE | -17.21 |
| Energy contribution | -17.34 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.977446 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
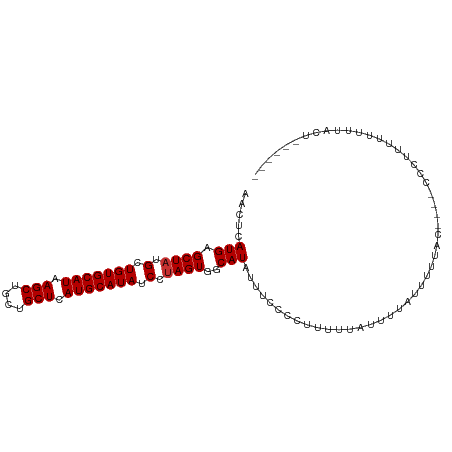
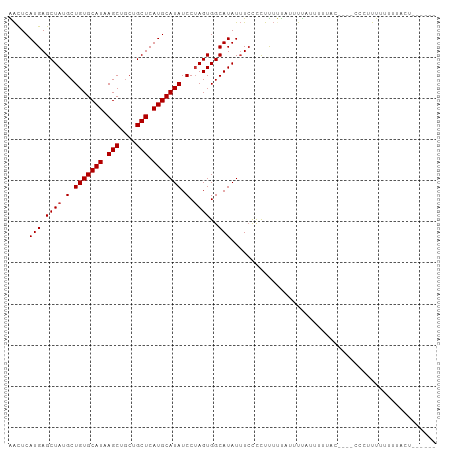
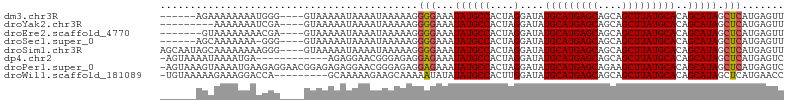
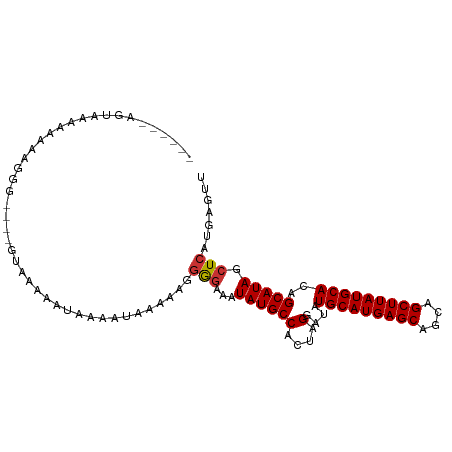
>dm3.chr3R 8771275 94 + 27905053 AACUCAUGAGCUAUGCUGUGCAUAAGCUGCUGCUCAUGCAUAUCCUAGUGGCAUAUUUCCCCUUUUUAUUUUAUUUUUAC----CCCAUUUUUUUUCU------ .....(((.((((.(.(((((((.(((....))).))))))).).))))..)))..........................----..............------ ( -17.50, z-score = -2.15, R) >droYak2.chr3R 13063468 91 + 28832112 AACUCAUGAGCUAUGCUGUGCAUAAGCUGCUGCUCAUGCAUAUCCUAGUGGCAUAUUUCCCCUUUUUAUUUUAUUUUUAC----UCGAUUUUUUU--------- .....(((.((((.(.(((((((.(((....))).))))))).).))))..)))..........................----...........--------- ( -17.50, z-score = -1.54, R) >droEre2.scaffold_4770 4866985 93 - 17746568 AACUCAUGAGCUAUGCUGUGCAUAAGCUGCUGCUCAUGCAUAUCCUAGUGGCAUAUUUCCCCUUUUUAUUUUAUUUUUAC----UCGUUUUUUUUAC------- .....(((.((((.(.(((((((.(((....))).))))))).).))))..)))..........................----.............------- ( -17.50, z-score = -1.73, R) >droSec1.super_0 7896134 93 - 21120651 AACUCAUGAGCUAUGCUGUGCAUAAGCUGCUGCUCAUGCAUAUCCUAGUGGCAUAUUUCCCCUUUUUAUUUUAUUUUUAC----CCC-UUUUUUUGCU------ .....(((.((((.(.(((((((.(((....))).))))))).).))))..)))..........................----...-..........------ ( -17.50, z-score = -1.62, R) >droSim1.chr3R 14866099 100 - 27517382 AACUCAUGAGCUAUGCUGUGCAUAAGCUGCUGCUCAUGCAUAUCCUAGUGGCAUAUUUCCCCUUUUUAUUUUAUUUUUAC----CCCUUUUUUUUGCUAUUGCU ....(((((((...((...((....)).)).))))))).......((((((((...........................----..........)))))))).. ( -18.69, z-score = -1.54, R) >dp4.chr2 19382004 91 - 30794189 GACUCAUGAGCUAUGCUGUGCAUAAGCUGCUGCUCAUGCAUAUCCUAGUGGCAUAUUUCUCCUCUCCCGUUCCUCU------------UCAUUUUAUUUUACU- .....(((.((((.(.(((((((.(((....))).))))))).).))))..)))......................------------...............- ( -17.50, z-score = -1.42, R) >droPer1.super_0 9105882 103 + 11822988 GACUCAUGAGCUAUGCUGUGCAUAAGCUUCUGCUCAUGCAUAUCCUAGUGGCAUAUUUCUCCUCUCCCGUUCCUCUCUCCGUUCCUCUUCAUUUUACUUUACU- .....(((.((((.(.(((((((.(((....))).))))))).).))))..))).................................................- ( -17.50, z-score = -2.24, R) >droWil1.scaffold_181089 11505755 94 + 12369635 GGUUCAUGAGCUAUGCUGUGCAUAAGCUGCUGCUCAUGCAUAUCCAAGUGGCAUAUAUAUUUUUGCUUCUUUUUGC---------UGGUCCUUUCUUUUUACA- ((..((...(((((..(((((((.(((....))).))))))).....)))))............((........))---------))..))............- ( -20.80, z-score = -0.10, R) >consensus AACUCAUGAGCUAUGCUGUGCAUAAGCUGCUGCUCAUGCAUAUCCUAGUGGCAUAUUUCCCCUUUUUAUUUUAUUUUUAC____CCCUUUUUUUUACU______ .....(((.((((.(.(((((((.(((....))).))))))).).))))..))).................................................. (-17.21 = -17.34 + 0.13)

| Location | 8,771,275 – 8,771,369 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 79.21 |
| Shannon entropy | 0.41111 |
| G+C content | 0.38542 |
| Mean single sequence MFE | -18.99 |
| Consensus MFE | -17.87 |
| Energy contribution | -17.89 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.936861 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 8771275 94 - 27905053 ------AGAAAAAAAAUGGG----GUAAAAAUAAAAUAAAAAGGGGAAAUAUGCCACUAGGAUAUGCAUGAGCAGCAGCUUAUGCACAGCAUAGCUCAUGAGUU ------.........(((((----.((..............((((........)).))......(((((((((....))))))))).....)).)))))..... ( -19.50, z-score = -1.56, R) >droYak2.chr3R 13063468 91 - 28832112 ---------AAAAAAAUCGA----GUAAAAAUAAAAUAAAAAGGGGAAAUAUGCCACUAGGAUAUGCAUGAGCAGCAGCUUAUGCACAGCAUAGCUCAUGAGUU ---------.........((----((...............((((........)).))......(((((((((....))))))))).......))))....... ( -18.80, z-score = -1.52, R) >droEre2.scaffold_4770 4866985 93 + 17746568 -------GUAAAAAAAACGA----GUAAAAAUAAAAUAAAAAGGGGAAAUAUGCCACUAGGAUAUGCAUGAGCAGCAGCUUAUGCACAGCAUAGCUCAUGAGUU -------...........((----((...............((((........)).))......(((((((((....))))))))).......))))....... ( -18.80, z-score = -1.63, R) >droSec1.super_0 7896134 93 + 21120651 ------AGCAAAAAAA-GGG----GUAAAAAUAAAAUAAAAAGGGGAAAUAUGCCACUAGGAUAUGCAUGAGCAGCAGCUUAUGCACAGCAUAGCUCAUGAGUU ------..........-...----...................(((...((((((....)....(((((((((....)))))))))..))))).)))....... ( -18.60, z-score = -0.83, R) >droSim1.chr3R 14866099 100 + 27517382 AGCAAUAGCAAAAAAAAGGG----GUAAAAAUAAAAUAAAAAGGGGAAAUAUGCCACUAGGAUAUGCAUGAGCAGCAGCUUAUGCACAGCAUAGCUCAUGAGUU .((....))...........----...................(((...((((((....)....(((((((((....)))))))))..))))).)))....... ( -19.80, z-score = -0.79, R) >dp4.chr2 19382004 91 + 30794189 -AGUAAAAUAAAAUGA------------AGAGGAACGGGAGAGGAGAAAUAUGCCACUAGGAUAUGCAUGAGCAGCAGCUUAUGCACAGCAUAGCUCAUGAGUC -...............------------...............(((...((((((....)....(((((((((....)))))))))..))))).)))....... ( -19.50, z-score = -1.27, R) >droPer1.super_0 9105882 103 - 11822988 -AGUAAAGUAAAAUGAAGAGGAACGGAGAGAGGAACGGGAGAGGAGAAAUAUGCCACUAGGAUAUGCAUGAGCAGAAGCUUAUGCACAGCAUAGCUCAUGAGUC -......................((..........))......(((...((((((....)....(((((((((....)))))))))..))))).)))....... ( -19.60, z-score = -1.43, R) >droWil1.scaffold_181089 11505755 94 - 12369635 -UGUAAAAAGAAAGGACCA---------GCAAAAAGAAGCAAAAAUAUAUAUGCCACUUGGAUAUGCAUGAGCAGCAGCUUAUGCACAGCAUAGCUCAUGAACC -..................---------.......((.((........((((.((....))))))((((((((....))))))))........))))....... ( -17.30, z-score = -0.09, R) >consensus ______AGUAAAAAAAAGGG____GUAAAAAUAAAAUAAAAAGGGGAAAUAUGCCACUAGGAUAUGCAUGAGCAGCAGCUUAUGCACAGCAUAGCUCAUGAGUU ...........................................(((...((((((....)....(((((((((....)))))))))..))))).)))....... (-17.87 = -17.89 + 0.02)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:10:39 2011