| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,743,705 – 8,743,810 |
| Length | 105 |
| Max. P | 0.997811 |

| Location | 8,743,705 – 8,743,810 |
|---|---|
| Length | 105 |
| Sequences | 9 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 79.28 |
| Shannon entropy | 0.44976 |
| G+C content | 0.31793 |
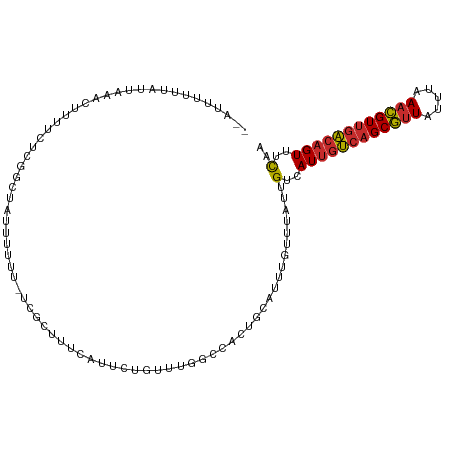
| Mean single sequence MFE | -20.76 |
| Consensus MFE | -12.70 |
| Energy contribution | -12.77 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.18 |
| SVM RNA-class probability | 0.997811 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
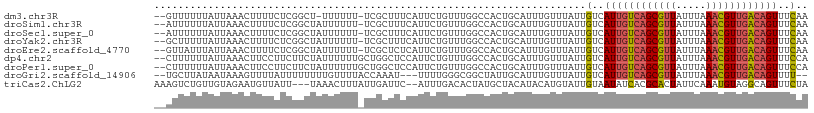
>dm3.chr3R 8743705 105 - 27905053 --GUUUUUUAUUAAACUUUUCUCGGCU-UUUUUU-UCGCUUUCAUUCUGUUUGGCCACUGCAUUUGUUUAUUGUCAUUGUCAGCGUUAUUUAAACGUUGACAGUUUCAA --((((......)))).......((((-......-.................))))..............(((..((((((((((((.....))))))))))))..))) ( -20.80, z-score = -3.29, R) >droSim1.chr3R 14844461 106 + 27517382 --AUUUUUUAUUAAACUUUUCUCGGCUAUUUUUU-UCGCUUUCAUUCUGUUUGGCCACUGCAUUUGUUUAUUGUCAUUGUCAGCGUUAUUUAAACGUUGACAGUUUCAA --.........(((((.......(((((......-................))))).........)))))(((..((((((((((((.....))))))))))))..))) ( -21.84, z-score = -3.83, R) >droSec1.super_0 7874621 106 + 21120651 --AUUUUUUAUUAAACUUUUCUCGGCUAUUUUUU-UCGCUUUCAUUCUGUUUGGCCACUGCAUUUGUUUAUUGUCAUUGUCAGCGUUAUUUAAACGUUGACAGUUUCAA --.........(((((.......(((((......-................))))).........)))))(((..((((((((((((.....))))))))))))..))) ( -21.84, z-score = -3.83, R) >droYak2.chr3R 13039729 106 - 28832112 --GCUUUUUAUUAAACUUUUCUCGGCUAUUUUUU-UCGCUUUCAUUCUGUUUGGCCACUGCAUUUGUUUAUUGUCAUUGUCAGCGUUAUUUAAACGUUGACAGUUUCAA --.........(((((.......(((((......-................))))).........)))))(((..((((((((((((.....))))))))))))..))) ( -21.84, z-score = -3.37, R) >droEre2.scaffold_4770 4843785 106 + 17746568 --GUUAUUUAUUAAACUUUUCUCGGCUAUUUUUU-UCGCUCUCAUUCUGUUUGGCCACUGCAUUUGUUUAUUGUCAUUGUCAGCGUUAUUUAAACGUUGACAGUUUCAA --.........(((((.......(((((......-................))))).........)))))(((..((((((((((((.....))))))))))))..))) ( -21.84, z-score = -3.60, R) >dp4.chr2 19354787 107 + 30794189 --CUUUUUUAUUAAACUUCCUUCUUCUAUUUUUUGCUGGCUCCAUUCUGUUUGGCCACUGCAUUUGUUUAUUGUCAUUGUCAGCGUUAUUUAAACGUUGACAGUUUCCA --...............................((((((((...........)))))..))).............((((((((((((.....))))))))))))..... ( -21.00, z-score = -3.37, R) >droPer1.super_0 9078539 107 - 11822988 --CUUUUUUAUUAAACUUCCUUCUUCUAUUUUUUGCUGGCUCCAUUCUGUUUGGCCACUGCAUUUGUUUAUUGUCAUUGUCAGCGUUAUUUAAACGUUGACAGUUUCCA --...............................((((((((...........)))))..))).............((((((((((((.....))))))))))))..... ( -21.00, z-score = -3.37, R) >droGri2.scaffold_14906 7473909 102 + 14172833 --UGCUUAUAAUAAAGUUUUAUUUUUUUUGUUUUACCAAAU---UUUUGGGCGGCUAUUGCAUUUGUUUAUUGUCAUUGUCAGCGUUAUUUAAACGUUGACAGUUUU-- --.....((((((((....................((((..---..))))...((....)).....)))))))).((((((((((((.....))))))))))))...-- ( -21.70, z-score = -2.37, R) >triCas2.ChLG2 1715497 104 - 12900155 AAAGUCUGUUGUAGAAUGUUAUU---UAAACUUUAUUGAUUC--AUUUGACACUAUGCUACAUACAUGUAUUGUAAUAUCACGCACUAUUCAAAUGUAGGCAGUUUCUA ..((.(((((((.(((((....(---(((......))))..)--)))).))).....((((((....(((.(((........))).)))....))))))))))...)). ( -15.00, z-score = 0.93, R) >consensus __AUUUUUUAUUAAACUUUUCUCGGCUAUUUUUU_UCGCUUUCAUUCUGUUUGGCCACUGCAUUUGUUUAUUGUCAUUGUCAGCGUUAUUUAAACGUUGACAGUUUCAA ........................................................................(..((((((((((((.....))))))))))))..).. (-12.70 = -12.77 + 0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:10:36 2011