| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,734,138 – 8,734,242 |
| Length | 104 |
| Max. P | 0.538530 |

| Location | 8,734,138 – 8,734,242 |
|---|---|
| Length | 104 |
| Sequences | 10 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 69.57 |
| Shannon entropy | 0.56716 |
| G+C content | 0.54653 |
| Mean single sequence MFE | -35.35 |
| Consensus MFE | -10.07 |
| Energy contribution | -10.30 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.61 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.538530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
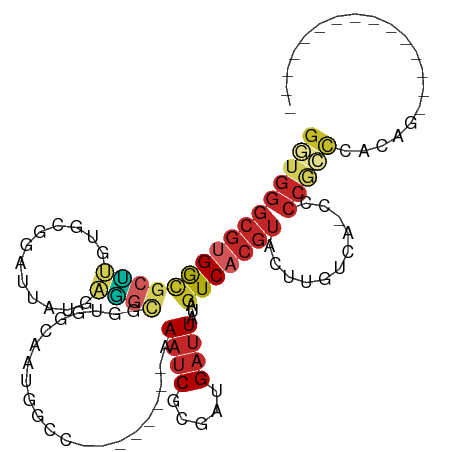
>dm3.chr3R 8734138 104 + 27905053 GUUGGGCGUGGCGCUUGUGCGGAUUAUGAAGCGGUGGCAAUGGCC--------AAAUCGCGAUGAUUAUAGUCACGUACUUGUCA-CUCCGCCCACAGCCCAUCCCCCAUCGC ..(((((((((.((..(((((((((((((.((((((((....)))--------)..)))).....)))))))).)))))..(...-...))))))).)))))........... ( -38.10, z-score = -1.20, R) >droEre2.scaffold_4770 4834343 90 - 17746568 GGUGGGCGUGGCACUUGUGCGGAUUAUGAGGCGGUGGCAAUGGCC--------AAAUCGCGAUGAUUAUAGUCACGUACUUGUCA-CCCCGCCCACAGU-------------- ((((((.((((((...(((((((((((((.((((((((....)))--------)..)))).....)))))))).))))).)))))-)))))))......-------------- ( -43.00, z-score = -3.67, R) >droYak2.chr3R 13030139 90 + 28832112 GGUGGGCGUGGCGCUUGUGCGGAUUAUGAGGCGGUGGCAAUGGCC--------AAAUCGCGAUGAUUAUAGUCACGUACUUGUCA-CCCCGCCCACAGC-------------- .(((((((.((.((..(((((((((((((.((((((((....)))--------)..)))).....)))))))).)))))..).).-)).)))))))...-------------- ( -42.80, z-score = -3.32, R) >droSec1.super_0 7865170 88 - 21120651 GGUGGGCGUGGCGCUUGUGCGGAUUAUGAAGCGGUGGCAAUGGCC--------AAAUCGCGAUGAUUAUAGUCACGUACUUGUCA-CCCCGCCCACA---------------- .(((((((.((.((..(((((((((((((.((((((((....)))--------)..)))).....)))))))).)))))..).).-)).))))))).---------------- ( -42.80, z-score = -3.79, R) >droSim1.chr3R 14834949 88 - 27517382 GGUGGGCGUGGCGCUUGUGCGGAUUAUGAAGCGGUGGCAAUGGCC--------AAAUCGCGAUGAUUAUAGUCACGUACUUGUCA-CCCCGCCCACA---------------- .(((((((.((.((..(((((((((((((.((((((((....)))--------)..)))).....)))))))).)))))..).).-)).))))))).---------------- ( -42.80, z-score = -3.79, R) >droWil1.scaffold_181089 11466511 78 + 12369635 ---------GGCGGCUGUGGGGAAUGGAAGAUUAUGA-GGCGGUA--------AAAUCGCGAUGAUUAUAGUCACGUACUUGUCA-C-CCACCCAAAC--------------- ---------...((..(((((((..(...((((((((-.(((((.--------..))))).....)))))))).....)...)).-)-))))))....--------------- ( -19.90, z-score = -0.26, R) >droPer1.super_0 9068275 88 + 11822988 GGUGGGCGAGAUGCUGUUGCAGAUUAUGAGGCGGCAAAAGCAA-----------AAUCGCGAUGAUUAUAGUCACGUACUUGCGC-CACCGCCUCUCUAU------------- ((((((((((.((((((..((.....))..)))))).......-----------....(((.((((....))))))).))))).)-))))..........------------- ( -32.00, z-score = -2.20, R) >dp4.chr2 19343572 88 - 30794189 GGUGGGCGUGAUGCUGUUGCAGAUUAUGAGGCGGCAAAAGCAA-----------AAUCGCGAUGAUUAUAGUCACGUACUUGCGC-CACCGCCUCUCUAU------------- ((((((((..(((((((..((.....))..)))))).......-----------....(((.((((....)))))))..)..)))-..))))).......------------- ( -28.90, z-score = -1.05, R) >droAna3.scaffold_13340 14264413 98 - 23697760 GGUGGGCGUGAAGCUUGUGCAGAUUAUGAGGCGGUGACGGUGGGUGGCGGUGGAAAUCGCGAUGAUUAUAGUCACGUACUUGUCA-CCCCAACUACCGU-------------- ((((((((..(...)..))).........((.((((((((((.((.(((((....)))))(((.......))))).))).)))))-))))..)))))..-------------- ( -37.60, z-score = -1.98, R) >droVir3.scaffold_12855 6070559 88 - 10161210 GAUGGGCGUGGUUUGGUUGCUGCUCUGACUCUGACAUUGCCGA-----------ACGCGUGUUGAUUAUAGUCACGUACUUGUCAACGCCCUUUGCAGC-------------- ...((((((.(((((((...((.((.......))))..)))))-----------))(((((...........)))))........))))))........-------------- ( -25.60, z-score = -0.33, R) >consensus GGUGGGCGUGGCGCUUGUGCGGAUUAUGAGGCGGUGGCAAUGGCC________AAAUCGCGAUGAUUAUAGUCACGUACUUGUCA_CCCCGCCCACAG_______________ (((((....(.(((((............))))).).......................(((.((((....)))))))...........))))).................... (-10.07 = -10.30 + 0.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:10:35 2011