| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,732,301 – 8,732,392 |
| Length | 91 |
| Max. P | 0.768624 |

| Location | 8,732,301 – 8,732,392 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 78.11 |
| Shannon entropy | 0.43194 |
| G+C content | 0.35479 |
| Mean single sequence MFE | -22.52 |
| Consensus MFE | -12.58 |
| Energy contribution | -12.26 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.610686 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
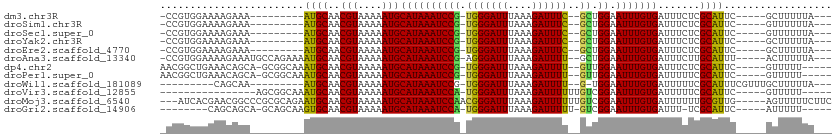
>dm3.chr3R 8732301 91 + 27905053 -CCGUGGAAAAGAAA---------AUGCAACGUAAAAAUGCAUAAAUCCG-UGGGAUUUAAAGAUUUC--GCUGGAAUUUGUGAUUUCUCGCAUUC-----GCUUUUUA--- -.....((((((..(---------((((..(((....)))((((((((((-((..(((....)))..)--)).)).))))))).......))))).-----.)))))).--- ( -23.90, z-score = -2.19, R) >droSim1.chr3R 14833097 91 - 27517382 -CCGUGGAAAAGAAA---------AUGCAACGUAAAAAUGCAUAAAUCCG-UGGGAUUUAAAGAUUUC--GCUGGAAUUUGUGAUUUCUCGCAUUC-----GUUUUUUA--- -.....((((((..(---------((((..(((....)))((((((((((-((..(((....)))..)--)).)).))))))).......))))).-----.)))))).--- ( -21.60, z-score = -1.50, R) >droSec1.super_0 7863312 91 - 21120651 -CCGUGGAAAAGAAA---------AUGCAACGUAAAAAUGCAUAAAUCCG-UGGGAUUUAAAGAUUUC--GCUGGAAUUUGUGAUUUCUCGCAUUC-----GUUUUUUA--- -.....((((((..(---------((((..(((....)))((((((((((-((..(((....)))..)--)).)).))))))).......))))).-----.)))))).--- ( -21.60, z-score = -1.50, R) >droYak2.chr3R 13028250 91 + 28832112 -CCGUGGAAAAGAAA---------AUGCAACGUAAAAAUGCAUAAAUCCG-UGGGAUUUAAAGAUUUC--GCUGGAAUUUGUGAUUUCUCGCAUUC-----GCUUUUUA--- -.....((((((..(---------((((..(((....)))((((((((((-((..(((....)))..)--)).)).))))))).......))))).-----.)))))).--- ( -23.90, z-score = -2.19, R) >droEre2.scaffold_4770 4832484 91 - 17746568 -CCGUGGAAAAGAAA---------AUGCAACGUAAAAAUGCAUAAAUCCG-UGGGAUUUAAAGAUUUC--GCUGGAAUUUGUGAUUUCUCGCAUUC-----GCUUUUUA--- -.....((((((..(---------((((..(((....)))((((((((((-((..(((....)))..)--)).)).))))))).......))))).-----.)))))).--- ( -23.90, z-score = -2.19, R) >droAna3.scaffold_13340 14262811 100 - 23697760 -CCGUGGAAAAGAAAUGCCAGAAAAUGCAACGUAAAAAUGCAUAAAUCCG-AGGGAUUUAAAGAUUUU--GCUGGAAUUUGUGAUUUCUUGCAUUU-----ACUUUUUA--- -.....((((((((((((.(((((.(((((.(((....))).....((((-.((((((....))))))--..))))..))))).))))).))))))-----.)))))).--- ( -23.50, z-score = -2.05, R) >dp4.chr2 11454182 98 + 30794189 AACGGCUGAAACAGCA-GCGGCAAAUGCAACGUAAAAAUGCAUAAAUCCG-UGGGAUUUAAAGAUUUU--GUUGGAAUUUGUGAUUUCUCGCAUUC-----GUUUUU----- ((((((((...)))).-(((((((((.(((((((....))).(((((((.-..)))))))........--))))..))))))(....).)))...)-----)))...----- ( -21.50, z-score = -0.07, R) >droPer1.super_0 2758394 98 - 11822988 AACGGCUGAAACAGCA-GCGGCAAAUGCAACGUAAAAAUGCAUAAAUCCG-UGGGAUUUAAAGAUUUU--GUUGGAAUUUGUGAUUUUUCGCAUUC-----GUUUUU----- ((((((((...)))).-(((((....)).....((((((.((((((((((-..(((((....))))).--.).)).))))))))))))))))...)-----)))...----- ( -23.00, z-score = -0.62, R) >droWil1.scaffold_181089 11463777 87 + 12369635 ---------CAGCAA---------AUGCAACGUAAAAAUGCAUAAAUCCG-UGGGAUUUAAAGAUUUU--G-UGGAAUUUGUGAUUUUUCGCAUUUCGUUUGCUUUUUA--- ---------.(((((---------(((.((.((((((((.((((((((((-..(((((....))))).--.-))).))))))))))))).)).)).)))))))).....--- ( -26.20, z-score = -3.70, R) >droVir3.scaffold_12855 6065294 85 - 10161210 ----------------AGCGGCAAAUGCAACGUAAAAAUGCAUAAAUCCA-UGGGAUUUAAAGAUUUUUUGUCGGAAUUUGUGAUUUUUCGCAUUC-----GUUUUU----- ----------------..(((((((((((.........))))(((((((.-..))))))).......)))))))(((..(((((....))))))))-----......----- ( -19.30, z-score = -1.20, R) >droMoj3.scaffold_6540 25729736 104 - 34148556 ---AUCACGAACGGCCCGCGCAGAAUGCAACGUAAAAAUGCAUAAAUCCAACGGGAUUUAAAGAUUUUUUGUCGGAAUUUGUGAUUUUUUGCGUUG-----AGUUUUUCUUC ---................(.(((((.(((((((((((..(((((((((.((((((..........)))))).)).)))))))..)))))))))))-----.))))).)... ( -25.40, z-score = -0.89, R) >droGri2.scaffold_14906 7463413 90 - 14172833 --------CAGCAGCA-GCAGCAAGUGCAACGUAAAAAUGCAUAAAUCCA-UGGGAUUUAAAGAUUUUU-GUCGGAAUUUGUGAUUU-UCGCAUUC-----AUUUUU----- --------..((....-...((((((....(((((((((...(((((((.-..)))))))...))))))-).))..)))))).....-..))....-----......----- ( -16.44, z-score = 0.33, R) >consensus _CCGUGGAAAAGAAA_________AUGCAACGUAAAAAUGCAUAAAUCCG_UGGGAUUUAAAGAUUUU__GCUGGAAUUUGUGAUUUCUCGCAUUC_____GUUUUUUA___ ....................................(((((.(((((((....))))))).(((((((.....)))))))..........)))))................. (-12.58 = -12.26 + -0.32)


| Location | 8,732,301 – 8,732,392 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.11 |
| Shannon entropy | 0.43194 |
| G+C content | 0.35479 |
| Mean single sequence MFE | -16.79 |
| Consensus MFE | -10.85 |
| Energy contribution | -10.75 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.768624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8732301 91 - 27905053 ---UAAAAAGC-----GAAUGCGAGAAAUCACAAAUUCCAGC--GAAAUCUUUAAAUCCCA-CGGAUUUAUGCAUUUUUACGUUGCAU---------UUUCUUUUCCACGG- ---..(((((.-----((((((((....)).........(((--((((....(((((((..-.)))))))......))).))))))))---------)).)))))......- ( -18.50, z-score = -2.04, R) >droSim1.chr3R 14833097 91 + 27517382 ---UAAAAAAC-----GAAUGCGAGAAAUCACAAAUUCCAGC--GAAAUCUUUAAAUCCCA-CGGAUUUAUGCAUUUUUACGUUGCAU---------UUUCUUUUCCACGG- ---..((((..-----((((((((....)).........(((--((((....(((((((..-.)))))))......))).))))))))---------))..))))......- ( -14.90, z-score = -1.15, R) >droSec1.super_0 7863312 91 + 21120651 ---UAAAAAAC-----GAAUGCGAGAAAUCACAAAUUCCAGC--GAAAUCUUUAAAUCCCA-CGGAUUUAUGCAUUUUUACGUUGCAU---------UUUCUUUUCCACGG- ---..((((..-----((((((((....)).........(((--((((....(((((((..-.)))))))......))).))))))))---------))..))))......- ( -14.90, z-score = -1.15, R) >droYak2.chr3R 13028250 91 - 28832112 ---UAAAAAGC-----GAAUGCGAGAAAUCACAAAUUCCAGC--GAAAUCUUUAAAUCCCA-CGGAUUUAUGCAUUUUUACGUUGCAU---------UUUCUUUUCCACGG- ---..(((((.-----((((((((....)).........(((--((((....(((((((..-.)))))))......))).))))))))---------)).)))))......- ( -18.50, z-score = -2.04, R) >droEre2.scaffold_4770 4832484 91 + 17746568 ---UAAAAAGC-----GAAUGCGAGAAAUCACAAAUUCCAGC--GAAAUCUUUAAAUCCCA-CGGAUUUAUGCAUUUUUACGUUGCAU---------UUUCUUUUCCACGG- ---..(((((.-----((((((((....)).........(((--((((....(((((((..-.)))))))......))).))))))))---------)).)))))......- ( -18.50, z-score = -2.04, R) >droAna3.scaffold_13340 14262811 100 + 23697760 ---UAAAAAGU-----AAAUGCAAGAAAUCACAAAUUCCAGC--AAAAUCUUUAAAUCCCU-CGGAUUUAUGCAUUUUUACGUUGCAUUUUCUGGCAUUUCUUUUCCACGG- ---..(((((.-----((((((.(((((..............--........(((((((..-.)))))))((((.........)))).))))).)))))))))))......- ( -19.80, z-score = -2.78, R) >dp4.chr2 11454182 98 - 30794189 -----AAAAAC-----GAAUGCGAGAAAUCACAAAUUCCAAC--AAAAUCUUUAAAUCCCA-CGGAUUUAUGCAUUUUUACGUUGCAUUUGCCGC-UGCUGUUUCAGCCGUU -----.....(-----((((((((....))............--(((((...(((((((..-.)))))))...)))))......)))))))..((-.((((...)))).)). ( -19.20, z-score = -1.24, R) >droPer1.super_0 2758394 98 + 11822988 -----AAAAAC-----GAAUGCGAAAAAUCACAAAUUCCAAC--AAAAUCUUUAAAUCCCA-CGGAUUUAUGCAUUUUUACGUUGCAUUUGCCGC-UGCUGUUUCAGCCGUU -----...(((-----(...(((((.................--........(((((((..-.)))))))((((.........))))))))).((-((......)))))))) ( -16.90, z-score = -0.68, R) >droWil1.scaffold_181089 11463777 87 - 12369635 ---UAAAAAGCAAACGAAAUGCGAAAAAUCACAAAUUCCA-C--AAAAUCUUUAAAUCCCA-CGGAUUUAUGCAUUUUUACGUUGCAU---------UUGCUG--------- ---......((((..(((((((((....))..........-.--........(((((((..-.))))))).)))))))....))))..---------......--------- ( -15.40, z-score = -2.16, R) >droVir3.scaffold_12855 6065294 85 + 10161210 -----AAAAAC-----GAAUGCGAAAAAUCACAAAUUCCGACAAAAAAUCUUUAAAUCCCA-UGGAUUUAUGCAUUUUUACGUUGCAUUUGCCGCU---------------- -----.....(-----((((((((....)).........(((.((((((...(((((((..-.)))))))...))))))..)))))))))).....---------------- ( -14.50, z-score = -1.35, R) >droMoj3.scaffold_6540 25729736 104 + 34148556 GAAGAAAAACU-----CAACGCAAAAAAUCACAAAUUCCGACAAAAAAUCUUUAAAUCCCGUUGGAUUUAUGCAUUUUUACGUUGCAUUCUGCGCGGGCCGUUCGUGAU--- ...........-----...........(((((.(((((((.(.((((((...(((((((....)))))))...)))))).....((.....)))))))..))).)))))--- ( -18.10, z-score = 0.29, R) >droGri2.scaffold_14906 7463413 90 + 14172833 -----AAAAAU-----GAAUGCGA-AAAUCACAAAUUCCGAC-AAAAAUCUUUAAAUCCCA-UGGAUUUAUGCAUUUUUACGUUGCACUUGCUGC-UGCUGCUG-------- -----......-----(((((((.-..............((.-.....))..(((((((..-.))))))))))))))....((.((((.....).-))).))..-------- ( -12.30, z-score = 0.12, R) >consensus ___UAAAAAAC_____GAAUGCGAGAAAUCACAAAUUCCAGC__AAAAUCUUUAAAUCCCA_CGGAUUUAUGCAUUUUUACGUUGCAU_________UUUCUUUUCCACGG_ ........(((.....((((((((....))......................(((((((....))))))).))))))....)))............................ (-10.85 = -10.75 + -0.10)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:10:34 2011