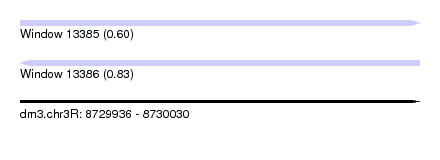
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,729,936 – 8,730,030 |
| Length | 94 |
| Max. P | 0.827482 |

| Location | 8,729,936 – 8,730,030 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 69.77 |
| Shannon entropy | 0.54793 |
| G+C content | 0.52917 |
| Mean single sequence MFE | -27.69 |
| Consensus MFE | -8.54 |
| Energy contribution | -13.16 |
| Covariance contribution | 4.62 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.604696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8729936 94 + 27905053 GUCGCUCCGGACAUCUGGUUCCCCAUGUACG-UAUGGAUGUAUCUACGUACUUACGUAUUUCCCAGAUCCAUUCACAACGCCCGUGAGUGCGACA (((((...(((..(((((........(((((-((...((((....))))...)))))))...))))))))((((((.......))))))))))). ( -31.30, z-score = -3.11, R) >droEre2.scaffold_4770 4830142 95 - 17746568 GUCGCUCCGGACAUCUGGUUCCCUAUGUACGCUAUGGAUGUAUCUACGUACUUACGUAUUUUCCAGAUCCAUUCACAACGCCCGUGAGCGCGACG (((((...(((..(((((.....((((((.(.((((..........))))).))))))....)))))))).(((((.......))))).))))). ( -28.00, z-score = -2.16, R) >droYak2.chr3R 13025949 94 + 28832112 GUCGCUCCGGACAUCUGGGUUCCCAUGUACG-UAUGGAUGUAUCUACGUACUUACGUAUUUCCCAGAUCCAUUCACAACGCCCGUGAGUGCGACG (((((...(((..((((((.......(((((-((...((((....))))...)))))))..)))))))))((((((.......))))))))))). ( -35.50, z-score = -3.80, R) >droSec1.super_0 7861016 94 - 21120651 GUCGCUCCGGACAUCUGGUUCCCCAUGUACG-UAUGGAUGUAUCUACGUACUUACGUAUUUCCCAGAUCCAUUCACAACGCCCGUGAGUGCGACG (((((...(((..(((((........(((((-((...((((....))))...)))))))...))))))))((((((.......))))))))))). ( -31.30, z-score = -3.07, R) >droSim1.chr3R 14830798 94 - 27517382 GUCGCUCCGGACAUCUGGUUCCCCAUGUUCG-UAUGGAUGUAUCUACGUACUUACGUAUUUCCCAGAUCCAUUCACAACGCCCGUGAGUGCGACG (((((...(((..(((((....(((((....-))))).......(((((....)))))....))))))))((((((.......))))))))))). ( -30.50, z-score = -3.01, R) >dp4.chr2 11452169 79 + 30794189 AAGGAUAUGGACAUUGGGUUCCACUCAGAAG------CCGGCCCCCCGUCCUCGCCUGUUCCGUACAUCUCUUGUAAAUGCGUCC---------- .(((....((((...((.(((......))).------))((....))))))...)))((....((((.....))))...))....---------- ( -18.60, z-score = 0.10, R) >droPer1.super_0 2756289 79 - 11822988 AAGGAUAUGGACAUUGGGUUCCACUCAGAAG------CCGGCCCCCCGUCCUCGCCUGUUCCGUACAUCUCUUGUAAAUGCGUCC---------- .(((....((((...((.(((......))).------))((....))))))...)))((....((((.....))))...))....---------- ( -18.60, z-score = 0.10, R) >consensus GUCGCUCCGGACAUCUGGUUCCCCAUGUACG_UAUGGAUGUAUCUACGUACUUACGUAUUUCCCAGAUCCAUUCACAACGCCCGUGAGUGCGACG (((((...(((..(((((....(((((.....))))).......(((((....)))))....)))))))).(((((.......))))).))))). ( -8.54 = -13.16 + 4.62)
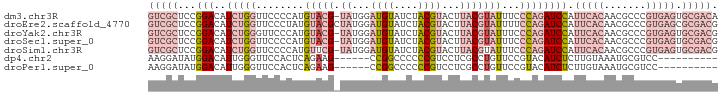
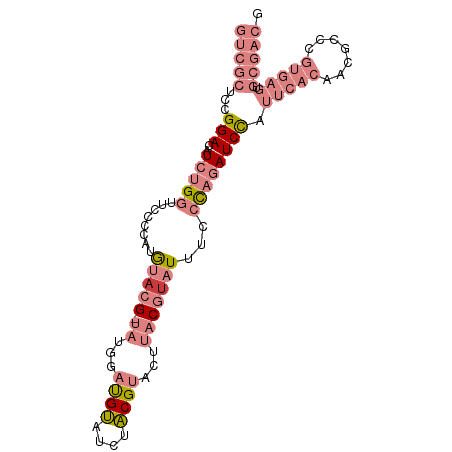
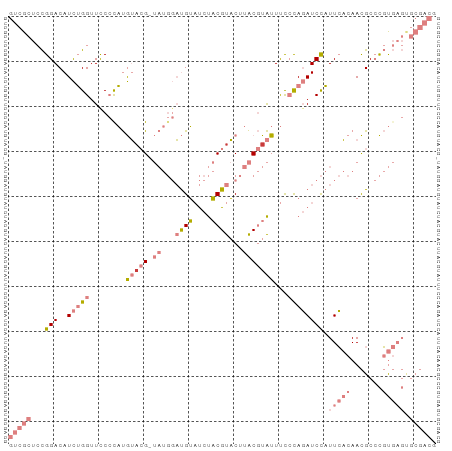
| Location | 8,729,936 – 8,730,030 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 69.77 |
| Shannon entropy | 0.54793 |
| G+C content | 0.52917 |
| Mean single sequence MFE | -31.01 |
| Consensus MFE | -11.70 |
| Energy contribution | -14.12 |
| Covariance contribution | 2.41 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.827482 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8729936 94 - 27905053 UGUCGCACUCACGGGCGUUGUGAAUGGAUCUGGGAAAUACGUAAGUACGUAGAUACAUCCAUA-CGUACAUGGGGAACCAGAUGUCCGGAGCGAC .(((((..((.(((((((((((.((((((.((.....(((((....)))))..)).)))))).-..))).(((....)))))))))))))))))) ( -34.30, z-score = -2.75, R) >droEre2.scaffold_4770 4830142 95 + 17746568 CGUCGCGCUCACGGGCGUUGUGAAUGGAUCUGGAAAAUACGUAAGUACGUAGAUACAUCCAUAGCGUACAUAGGGAACCAGAUGUCCGGAGCGAC .....(((((.(((((((((((.(((.(((((.....(((....)))..))))).))).))))))))((((..(....)..)))))))))))).. ( -33.80, z-score = -2.73, R) >droYak2.chr3R 13025949 94 - 28832112 CGUCGCACUCACGGGCGUUGUGAAUGGAUCUGGGAAAUACGUAAGUACGUAGAUACAUCCAUA-CGUACAUGGGAACCCAGAUGUCCGGAGCGAC .(((((..((.(((((((((((.((((((.((.....(((((....)))))..)).)))))).-..))).(((....)))))))))))))))))) ( -33.60, z-score = -2.29, R) >droSec1.super_0 7861016 94 + 21120651 CGUCGCACUCACGGGCGUUGUGAAUGGAUCUGGGAAAUACGUAAGUACGUAGAUACAUCCAUA-CGUACAUGGGGAACCAGAUGUCCGGAGCGAC .(((((..((.(((((((((((.((((((.((.....(((((....)))))..)).)))))).-..))).(((....)))))))))))))))))) ( -34.60, z-score = -2.76, R) >droSim1.chr3R 14830798 94 + 27517382 CGUCGCACUCACGGGCGUUGUGAAUGGAUCUGGGAAAUACGUAAGUACGUAGAUACAUCCAUA-CGAACAUGGGGAACCAGAUGUCCGGAGCGAC .(((((..((.(((((((((((.(((.(((((.(...(((....)))).))))).))).))).-......(((....)))))))))))))))))) ( -34.20, z-score = -2.94, R) >dp4.chr2 11452169 79 - 30794189 ----------GGACGCAUUUACAAGAGAUGUACGGAACAGGCGAGGACGGGGGGCCGG------CUUCUGAGUGGAACCCAAUGUCCAUAUCCUU ----------((((((...((((.....))))(......)))).((((.(((..(((.------((....)))))..)))...))))...))).. ( -23.30, z-score = -0.35, R) >droPer1.super_0 2756289 79 + 11822988 ----------GGACGCAUUUACAAGAGAUGUACGGAACAGGCGAGGACGGGGGGCCGG------CUUCUGAGUGGAACCCAAUGUCCAUAUCCUU ----------((((((...((((.....))))(......)))).((((.(((..(((.------((....)))))..)))...))))...))).. ( -23.30, z-score = -0.35, R) >consensus CGUCGCACUCACGGGCGUUGUGAAUGGAUCUGGGAAAUACGUAAGUACGUAGAUACAUCCAUA_CGUACAUGGGGAACCAGAUGUCCGGAGCGAC .(((((.....((((((((.........................(((((...............))))).(((....)))))))))))..))))) (-11.70 = -14.12 + 2.41)

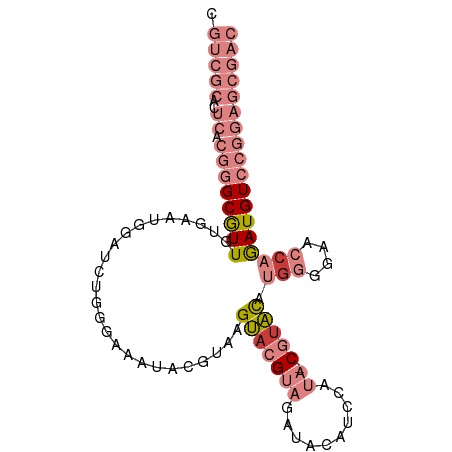
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:10:32 2011