
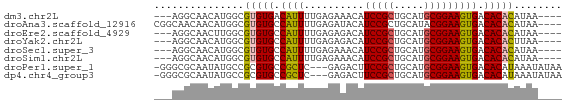
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,243,569 – 7,243,630 |
| Length | 61 |
| Max. P | 0.786507 |

| Location | 7,243,569 – 7,243,630 |
|---|---|
| Length | 61 |
| Sequences | 8 |
| Columns | 68 |
| Reading direction | forward |
| Mean pairwise identity | 81.72 |
| Shannon entropy | 0.33772 |
| G+C content | 0.50669 |
| Mean single sequence MFE | -18.20 |
| Consensus MFE | -14.73 |
| Energy contribution | -14.54 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.21 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.544319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 7243569 61 + 23011544 ---AGGCAACAUGGCGUGUGACAUUUUGAGAAACAUCCGCUGCAUGCGGAAGUGACACACAUAA---- ---.(....).....(((((.((((..........(((((.....))))))))).)))))....---- ( -17.10, z-score = -1.34, R) >droAna3.scaffold_12916 15145267 64 + 16180835 CGGCAACAACAUGGCGUGUGCCAUUUUGAGAUACAUCCGCUGCAUACGGAAGUGACACACAUAA---- ((((..(((.(((((....))))).))).((....)).))))..(((....)))..........---- ( -14.50, z-score = -0.18, R) >droEre2.scaffold_4929 16163234 61 + 26641161 ---AGGCAACUUGGCGUGUGCCAUUUUGAGAGACAUCCGCUGCAUGCGGAAGUGACACACAUAA---- ---..((......))(((((.((((((....))..(((((.....))))))))).)))))....---- ( -18.40, z-score = -1.16, R) >droYak2.chr2L 16664954 61 - 22324452 ---AGGCAACAUGGCGUGUGCCAUUUUGAGAGACAUCCGCUGCAUGCGGAAGUGACACACUUAA---- ---.(....).....(((((.((((((....))..(((((.....))))))))).)))))....---- ( -18.00, z-score = -1.01, R) >droSec1.super_3 2766303 61 + 7220098 ---AGGCAACAUGGCGUGUGCCAUUUUGAGAAACAUCCGCUGCAUGCGGAAGUGACACACAUAA---- ---...(((.(((((....))))).))).(..((.(((((.....))))).))..)........---- ( -17.80, z-score = -1.02, R) >droSim1.chr2L 7033888 61 + 22036055 ---AGGCAACAUGGCGUGUGCCAUUUUGAGAAACAUCCGCUGCAUGCGGAAGUGACACACAUAA---- ---...(((.(((((....))))).))).(..((.(((((.....))))).))..)........---- ( -17.80, z-score = -1.02, R) >droPer1.super_1 7940015 64 - 10282868 -GGGCGCAAUAUGCCGCGUGCCGCUC---GAGACUUCCGCUGCAUGCGGAAGUGACACAUAAAUAUAA -.(((((..........)))))....---...((((((((.....))))))))............... ( -21.00, z-score = -1.06, R) >dp4.chr4_group3 10829303 64 - 11692001 -GGGCGCAAUAUGCCGCGUGCCGCUC---GAGACUUCCGCUGCAUGCGGAAGUGACACAUAAAUAUAA -.(((((..........)))))....---...((((((((.....))))))))............... ( -21.00, z-score = -1.06, R) >consensus ___AGGCAACAUGGCGUGUGCCAUUUUGAGAGACAUCCGCUGCAUGCGGAAGUGACACACAUAA____ ...............(((((.((((..........(((((.....))))))))).)))))........ (-14.73 = -14.54 + -0.19)


| Location | 7,243,569 – 7,243,630 |
|---|---|
| Length | 61 |
| Sequences | 8 |
| Columns | 68 |
| Reading direction | reverse |
| Mean pairwise identity | 81.72 |
| Shannon entropy | 0.33772 |
| G+C content | 0.50669 |
| Mean single sequence MFE | -18.50 |
| Consensus MFE | -15.33 |
| Energy contribution | -15.41 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.786507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 7243569 61 - 23011544 ----UUAUGUGUGUCACUUCCGCAUGCAGCGGAUGUUUCUCAAAAUGUCACACGCCAUGUUGCCU--- ----....(((((.((..(((((.....)))))((.....))...)).)))))............--- ( -15.10, z-score = -0.86, R) >droAna3.scaffold_12916 15145267 64 - 16180835 ----UUAUGUGUGUCACUUCCGUAUGCAGCGGAUGUAUCUCAAAAUGGCACACGCCAUGUUGUUGCCG ----....((((((((..(((((.....)))))((.....))...))))))))............... ( -16.50, z-score = -0.38, R) >droEre2.scaffold_4929 16163234 61 - 26641161 ----UUAUGUGUGUCACUUCCGCAUGCAGCGGAUGUCUCUCAAAAUGGCACACGCCAAGUUGCCU--- ----....((((((((..(((((.....)))))((.....))...))))))))............--- ( -18.40, z-score = -1.64, R) >droYak2.chr2L 16664954 61 + 22324452 ----UUAAGUGUGUCACUUCCGCAUGCAGCGGAUGUCUCUCAAAAUGGCACACGCCAUGUUGCCU--- ----....((((((((..(((((.....)))))((.....))...))))))))............--- ( -18.60, z-score = -1.72, R) >droSec1.super_3 2766303 61 - 7220098 ----UUAUGUGUGUCACUUCCGCAUGCAGCGGAUGUUUCUCAAAAUGGCACACGCCAUGUUGCCU--- ----....((((((((..(((((.....)))))((.....))...))))))))............--- ( -18.40, z-score = -1.40, R) >droSim1.chr2L 7033888 61 - 22036055 ----UUAUGUGUGUCACUUCCGCAUGCAGCGGAUGUUUCUCAAAAUGGCACACGCCAUGUUGCCU--- ----....((((((((..(((((.....)))))((.....))...))))))))............--- ( -18.40, z-score = -1.40, R) >droPer1.super_1 7940015 64 + 10282868 UUAUAUUUAUGUGUCACUUCCGCAUGCAGCGGAAGUCUC---GAGCGGCACGCGGCAUAUUGCGCCC- ........(((((((((((((((.....))))))))...---..(((...)))))))))).......- ( -21.30, z-score = -1.22, R) >dp4.chr4_group3 10829303 64 + 11692001 UUAUAUUUAUGUGUCACUUCCGCAUGCAGCGGAAGUCUC---GAGCGGCACGCGGCAUAUUGCGCCC- ........(((((((((((((((.....))))))))...---..(((...)))))))))).......- ( -21.30, z-score = -1.22, R) >consensus ____UUAUGUGUGUCACUUCCGCAUGCAGCGGAUGUCUCUCAAAAUGGCACACGCCAUGUUGCCU___ ........(((((((((.(((((.....))))).))..........)))))))............... (-15.33 = -15.41 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:23:17 2011