| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,675,873 – 8,675,961 |
| Length | 88 |
| Max. P | 0.996575 |

| Location | 8,675,873 – 8,675,961 |
|---|---|
| Length | 88 |
| Sequences | 7 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 62.56 |
| Shannon entropy | 0.74329 |
| G+C content | 0.37380 |
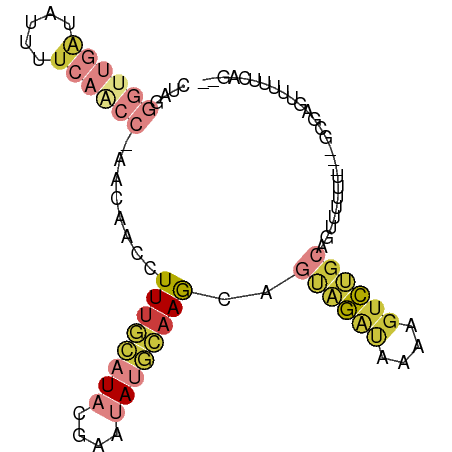
| Mean single sequence MFE | -21.22 |
| Consensus MFE | -8.32 |
| Energy contribution | -10.04 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.996575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
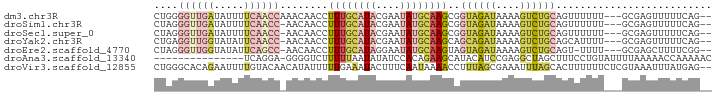
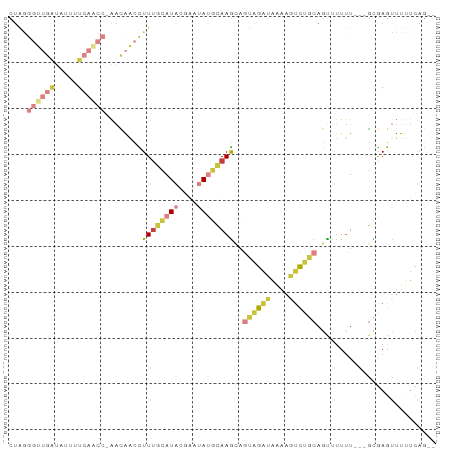
>dm3.chr3R 8675873 88 - 27905053 CUGGGGUUGAUAUUUUCAACCAAACAACCUUUGCAUACGAAUAUGCAAGCGGUAGAUAAAAGUCUGCAGUUUUUU---GCGAGUUUUUCAG-- .((.((((((.....))))))...))(((((((((((....)))))))).))).((.((((.((.((((....))---)))).))))))..-- ( -25.80, z-score = -3.24, R) >droSim1.chr3R 14776410 87 + 27517382 CUAGGGUUGAUAUUUUCAACC-AACAACCUUUGCAUACGAAUAUGCAAGCGGUAGAUAAAAGUCUGCAGUUUUUU---GCGAGUUUUUCAG-- ....((((((.....))))))-....(((((((((((....)))))))).))).((.((((.((.((((....))---)))).))))))..-- ( -25.50, z-score = -3.63, R) >droSec1.super_0 7807159 87 + 21120651 CUAGGGUUGAUAUUUUCAACC-AACAACCUUUGCAUACGAAUAUGCAAGCGGUAGAUAAAAGUCUGCAGUUUUUU---GCGAGUUUUUCAG-- ....((((((.....))))))-....(((((((((((....)))))))).))).((.((((.((.((((....))---)))).))))))..-- ( -25.50, z-score = -3.63, R) >droYak2.chr3R 12969701 87 - 28832112 CUGAGGUUGGUAUAUUCAACC-AACAACCUUUGCAUACGAAUAUGCAAGCAGCAGAUAAAAGUCUGCAGCAUUUU---GCGAGUUUUUCAG-- ((((((((((.....))))))-(((.....(((((((....)))))))((.((((((....)))))).)).....---....)))..))))-- ( -26.60, z-score = -3.48, R) >droEre2.scaffold_4770 4773503 86 + 17746568 CUAGGGUUGGUAUAUUCAGCC-AACAACCUUUGCAUAGGAAUAUGCAAGUAGUAGAUAAAAGUCUGCAGU-UUUU---GCGAGCUUUUCGG-- .....((((((.......)))-)))..((((((((((....))))))))........(((((..(((((.-..))---)))..))))).))-- ( -22.30, z-score = -1.53, R) >droAna3.scaffold_13340 14210763 77 + 23697760 ---------------UCAGGA-GGGGUCUUUUUAAUAUAUCCACAGAAGCAUACAUCCGAGGCUAGCUUUCCUGUAUUUUAAAAACCAAAAAC ---------------.(((((-((((((((............................)))))...))))))))................... ( -10.49, z-score = 0.50, R) >droVir3.scaffold_12855 5995671 91 + 10161210 CUGGGCACAGAAUUUUGUACAACAUAUUUUUGAAAUACUUUCAAUAAAACCUUUAGCGAAAUUUAGCACUUUUUUCUCGUAAAUUUAUGAG-- ....((...(((((((((...........(((((.....)))))...........))))))))).))........((((((....))))))-- ( -12.35, z-score = -0.87, R) >consensus CUAGGGUUGAUAUUUUCAACC_AACAACCUUUGCAUACGAAUAUGCAAGCAGUAGAUAAAAGUCUGCAGUUUUUU___GCGAGUUUUUCAG__ ....((((((.....))))))........((((((((....))))))))..((((((....)))))).......................... ( -8.32 = -10.04 + 1.72)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:10:25 2011