
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,672,158 – 8,672,273 |
| Length | 115 |
| Max. P | 0.827580 |

| Location | 8,672,158 – 8,672,273 |
|---|---|
| Length | 115 |
| Sequences | 9 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 75.99 |
| Shannon entropy | 0.48356 |
| G+C content | 0.44806 |
| Mean single sequence MFE | -27.86 |
| Consensus MFE | -16.38 |
| Energy contribution | -16.92 |
| Covariance contribution | 0.54 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.827580 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8672158 115 - 27905053 AGUAGCAGCACAUCUUGAGUGCAGUUUAAACUUAAAUUGUAUUUUUGAAAUGCUCGCAAUUAGGCAAACUGCCAAAGGCAGC-CGCCCUCCGCAUUUACCCGCUUUUCCACGACUU ....((((((..((..((((((((((((....))))))))))))..))..)))).)).....(((...(((((...))))).-.)))....((........))............. ( -30.90, z-score = -2.23, R) >droSim1.chr3R 14772710 115 + 27517382 AGUAGCUGCACAUCUUGAGUGCAGUUUAAACUUAAAUUGUAUUUUUGAAAUGCUCGCAAUUAGGCAAACUGCCAAAGGCAGC-CGCCCGCCGCAUUUUCCCGCUUUUCCACGACUU .(..((.(((..((..((((((((((((....))))))))))))..))..))).........(((...(((((...))))).-.))).))..)....................... ( -29.70, z-score = -1.25, R) >droSec1.super_0 7803487 115 + 21120651 AGUAGCUGCAAAUCGUGAGUGCAGUUUAAACUUAAAUUGUAUUUUUGAAAUGCUCGCAAUUAGGCAAACUGCCAAAGGCAUC-CGCCCGCCGCAUUUUCCCGCUUUUCCACAACUU ...(((.(((..(((.((((((((((((....)))))))))))).)))..)))..((.....(((.....)))...(((...-.....)))))........)))............ ( -29.00, z-score = -1.57, R) >droYak2.chr3R 12965948 105 - 28832112 ----------CAUCUUGAGUGCAGUUUAAACUUAAAUUGUAUUUUUGAAAUGCUCGCAAUUAGGCAAACUGCCAAAGGCAGC-CGCCCGCCGCAUUUUCCCGCUUUCCCACGACUU ----------..((..((((((((((((....))))))))))))..))(((((..((.....(((.....)))...(((...-.))).)).))))).................... ( -26.10, z-score = -1.63, R) >droEre2.scaffold_4770 4769782 115 + 17746568 AGUAGCAGCACAUCUUGAGUGCAGUUUAAACUUAAAUUGUAUUUUUGAAAUGCUCGCAAUUAGGCAAACUGCCAAAGGCAGC-CGCCCGCCGCAUUUUCCCGCUUUUCCACGACUU .(..((((((..((..((((((((((((....))))))))))))..))..))))........(((...(((((...))))).-.))).))..)....................... ( -31.00, z-score = -1.77, R) >droAna3.scaffold_13340 14206480 113 + 23697760 AACUGCAGCACAUCUUGAGUGCAGUUUAAACUUAAAUUGUAUUUUUGAAAUGUUCGCAAUUAGGCAGACUGCCAAAGGCAUC-CCUCCAUUUCUCCUGCUCUUGCUCACUCCUU-- ...(((((((..((..((((((((((((....))))))))))))..))..)))).)))...(((.((..((((...))))..-..............((....))...))))).-- ( -24.90, z-score = -1.17, R) >droVir3.scaffold_12855 5992000 95 + 10161210 UGCUACUGCACAGCUUGAGUGCAGUUUAAAGUUAAAUUGUAUUUCGGAAAUGUUCGCAAUUAGGCAGCCUGCCAAAGGCAUCUCCUCCAUCGCCU--------------------- ((((..((((((.((.((((((((((((....)))))))))))).))...)))..)))....))))((((.....))))................--------------------- ( -24.40, z-score = -0.72, R) >droMoj3.scaffold_6540 25638234 95 + 34148556 UGCUGCUGCACAGCUUGAGUGCAGUUUAAAGUUAAAUUGUAUUUCGGAAAUGUUCGCAAUUAGGCAGCCUGCCAAAGGCAUCGCCUCCAUCGCCU--------------------- .(((((.(.(((.((.((((((((((((....)))))))))))).))...))).))))...((((.((((.....))))...)))).....))..--------------------- ( -27.20, z-score = -0.61, R) >droGri2.scaffold_14906 7401135 91 + 14172833 UGUUGCACAGCAGCUUGAGUGCAGUUU-AAGUUAAAUUGUAUUUCGGAAAUGUUUGCAAUUAGGCAGCCUGCCAAAGGCAA---CCCCAUCGCCU--------------------- .((((((.((((.((.(((((((((((-(...)))))))))))).))...))))))))))..(((.....)))..((((..---.......))))--------------------- ( -27.50, z-score = -1.59, R) >consensus AGUAGCAGCACAUCUUGAGUGCAGUUUAAACUUAAAUUGUAUUUUUGAAAUGCUCGCAAUUAGGCAAACUGCCAAAGGCAGC_CGCCCACCGCAUUUUCCCGCUUUUCCACGACUU .......(((...(..((((((((((((....))))))))))))..)...)))..((.....(((.....)))...((........))...))....................... (-16.38 = -16.92 + 0.54)

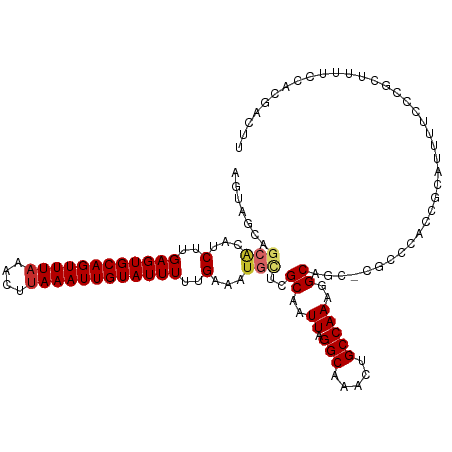
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:10:23 2011