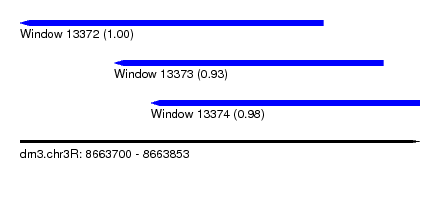
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,663,700 – 8,663,853 |
| Length | 153 |
| Max. P | 0.996302 |

| Location | 8,663,700 – 8,663,816 |
|---|---|
| Length | 116 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.37 |
| Shannon entropy | 0.07678 |
| G+C content | 0.35179 |
| Mean single sequence MFE | -32.19 |
| Consensus MFE | -28.29 |
| Energy contribution | -28.30 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.65 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.91 |
| SVM RNA-class probability | 0.996302 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8663700 116 - 27905053 AAACGUAAACUCUUCAAUAAUUGAACGUAUAGUCAUGAGGAAGCGAUAAAAUCGAAUUGCUUCCUUAAUGUGUGAAAUUGAUACAGGAAACAAUUUUCCACAUUGUUAUGGCUCGC---- ..((((.......(((.....)))))))..(((((((((((((((((........))))))))).(((((((..((((((..........))))))..)))))))))))))))...---- ( -31.51, z-score = -3.72, R) >droPer1.super_0 2829889 115 + 11822988 -AACGUAAACUCUUCAAUAAUUGAACGUGUAGUCAUGAGGAAGCGAUAAAAUCGAAUUGCUUCCUUAAUGUGUGAAAUUGAUACAGGAAACAAUUUUCCACAUUGUUAUGGCUCGC---- -...........((((.....)))).(((.(((((((((((((((((........))))))))).(((((((..((((((..........))))))..))))))))))))))))))---- ( -33.80, z-score = -4.10, R) >dp4.chr2 11525841 115 - 30794189 -AACGUAAACUCUUCAAUAAUUGAACGUGUAGUCAUGAGGAAGCGAUAAAAUCGAAUUGCUUCCUUAAUGUGUGAAAUUGAUACAGGAAACAAUUUUCCACAUUGUUAUGGCUCGC---- -...........((((.....)))).(((.(((((((((((((((((........))))))))).(((((((..((((((..........))))))..))))))))))))))))))---- ( -33.80, z-score = -4.10, R) >droAna3.scaffold_13340 14198671 116 + 23697760 AAACGUAAACUCUUCAAUAAUUGAACGUGUAGUCAUGAGGAAGCGAUAAAAUCGAAUUGCUUCCUUAAUGUGUGAAAUUGAUACAGGAAACAAUUUUCCACAUUGUUAUGGCCCGC---- ........((.(((((.....)))).).)).((((((((((((((((........))))))))).(((((((..((((((..........))))))..))))))))))))))....---- ( -31.10, z-score = -3.32, R) >droEre2.scaffold_4770 4761381 116 + 17746568 AAACGUAAACUCUUCAAUAAUUGAACGUGUAGUCAUGAGGAAGCGAUAAAAUCGAAUUGCUUCCUUAAUGUGUGAAAUUGAUACAGGAAACAAUUUUCCACAUUGUUAUGGCUCGC---- ............((((.....)))).(((.(((((((((((((((((........))))))))).(((((((..((((((..........))))))..))))))))))))))))))---- ( -33.80, z-score = -4.14, R) >droYak2.chr3R 12957313 116 - 28832112 AAACGUAAACUCUUCAAUAAUUGAACGUGUAGUCAUGAGGAAGCGAUAAAAUCGAAUUGCUUCCUUAAUGUGUGAAAUUGAUACAGGAAACAAUUUUCCACAUUGUUAUGGCUCGC---- ............((((.....)))).(((.(((((((((((((((((........))))))))).(((((((..((((((..........))))))..))))))))))))))))))---- ( -33.80, z-score = -4.14, R) >droSec1.super_0 7794962 116 + 21120651 AAACGUAAACUCUUCAAUAAUUGAACGUGUAGUCAUGAGGAAGCGAUAAAAUCGAAUUGCUUCCUUAAUGUGUGAAAUUGAUACAGGAAACAAUUUUCCACAUUGUUAUGGCUCGC---- ............((((.....)))).(((.(((((((((((((((((........))))))))).(((((((..((((((..........))))))..))))))))))))))))))---- ( -33.80, z-score = -4.14, R) >droSim1.chr3R 14764047 116 + 27517382 AAACGUAAACUCUUCAAUAAUUGAACGUGUAGUCAUGAGGAAGCGAUAAAAUCGAAUUGCUUCCUUAAUGUGUGAAAUUGAUACAGGAAACAAUUUUCCACAUUGUUAUGGCUCGC---- ............((((.....)))).(((.(((((((((((((((((........))))))))).(((((((..((((((..........))))))..))))))))))))))))))---- ( -33.80, z-score = -4.14, R) >droVir3.scaffold_12855 5981984 115 + 10161210 -AACGUAAACUCUUCAAUAAUUGAACGUGAAGUCAUGAGGAAGCGAUAAAAUCAAAUUGUUUCCUUAAUGUGUGAAAUUGAUACAGGAAACAAUUUUCCACAUUGUUAUGAGGCGC---- -..(((.....(((((...........)))))(((((((((((((((........))))))))).(((((((..((((((..........))))))..))))))))))))).))).---- ( -28.90, z-score = -2.71, R) >droMoj3.scaffold_6540 25625319 115 + 34148556 -AACGUAAACUCUUCAAUAAUUGAACGUGAAGUCAUGAGGAAGCGAUAAAAUCAAAUUGCUUCCUUAAUGUGUGAAAUUGAUACAGGAAACAAUUUUCCACAUUGUUAUGAGGCGC---- -..(((.....(((((...........)))))(((((((((((((((........))))))))).(((((((..((((((..........))))))..))))))))))))).))).---- ( -31.30, z-score = -3.49, R) >droGri2.scaffold_14906 7392799 115 + 14172833 -AACGUAAACUCUUCAAUAAUUGAACGUGAAGUCAUGAGGAAGCGAUAAAAUCAAAUUGCUUCCUUAAUGUGUGAAAUUGAUACAGGAAACAAUUUUCCACAUUGUUAUGAGGCGC---- -..(((.....(((((...........)))))(((((((((((((((........))))))))).(((((((..((((((..........))))))..))))))))))))).))).---- ( -31.30, z-score = -3.49, R) >droWil1.scaffold_181089 11381097 119 - 12369635 -AACGUAAACUCUUCAAUAAUUGAACGUGUAGUCAUGAGGAAGCGAUAAAAUCGAAUUGCUUCCUUAAUGUGUGAAAUUGAUACAGGAAACAAUUUUCCACAUUGUUAUGGGAUGGAAGC -..........((((........(((..........(((((((((((........)))))))))))((((((..((((((..........))))))..)))))))))........)))). ( -29.39, z-score = -2.25, R) >consensus _AACGUAAACUCUUCAAUAAUUGAACGUGUAGUCAUGAGGAAGCGAUAAAAUCGAAUUGCUUCCUUAAUGUGUGAAAUUGAUACAGGAAACAAUUUUCCACAUUGUUAUGGCUCGC____ ...(((((.................((((....))))((((((((((........))))))))))(((((((..((((((..........))))))..)))))))))))).......... (-28.29 = -28.30 + 0.01)
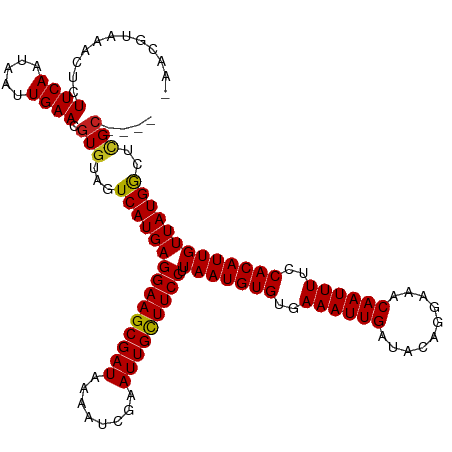
| Location | 8,663,736 – 8,663,839 |
|---|---|
| Length | 103 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 85.54 |
| Shannon entropy | 0.31441 |
| G+C content | 0.40347 |
| Mean single sequence MFE | -26.32 |
| Consensus MFE | -19.69 |
| Energy contribution | -19.72 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.926816 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
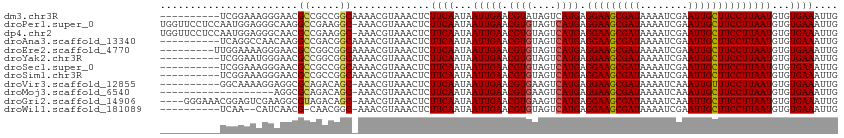
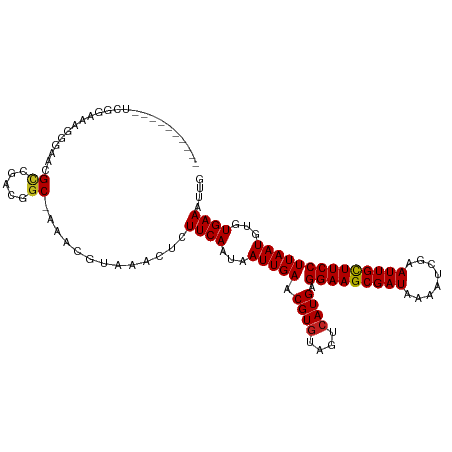
>dm3.chr3R 8663736 103 - 27905053 ----------UCGGAAAGGGAACGCCGCCGGCAAAACGUAAACUCUUCAAUAAUUGAACGUAUAGUCAUGAGGAAGCGAUAAAAUCGAAUUGCUUCCUUAAUGUGUGAAAUUG ----------.(((...((.....)).)))(((..((((.......(((.....))))))).......((((((((((((........)))))))))))).)))......... ( -23.61, z-score = -1.11, R) >droPer1.super_0 2829925 112 + 11822988 UGGUUCCUCCAAUGGAGGGCAAGGCCGAAGGC-AAACGUAAACUCUUCAAUAAUUGAACGUGUAGUCAUGAGGAAGCGAUAAAAUCGAAUUGCUUCCUUAAUGUGUGAAAUUG .(((((((((...))))))....)))(((((.-..........)))))..(((((..(((..(.....((((((((((((........)))))))))))))..)))..))))) ( -31.20, z-score = -2.21, R) >dp4.chr2 11525877 112 - 30794189 UGGUUCCUCCAAUGGAGGGCAACGCCGAAGGC-AAACGUAAACUCUUCAAUAAUUGAACGUGUAGUCAUGAGGAAGCGAUAAAAUCGAAUUGCUUCCUUAAUGUGUGAAAUUG .(((((((((...))))))....)))(((((.-..........)))))..(((((..(((..(.....((((((((((((........)))))))))))))..)))..))))) ( -31.20, z-score = -2.34, R) >droAna3.scaffold_13340 14198707 103 + 23697760 ----------UCAGGCCAACAAGGCCGACGGCAAAACGUAAACUCUUCAAUAAUUGAACGUGUAGUCAUGAGGAAGCGAUAAAAUCGAAUUGCUUCCUUAAUGUGUGAAAUUG ----------...((((.....)))).(((......)))......((((...(((((.((((....)))).(((((((((........))))))))))))))...)))).... ( -29.30, z-score = -3.08, R) >droEre2.scaffold_4770 4761417 104 + 17746568 ---------UUGGAAAAGGGAACGCCGGCGGCAAAACGUAAACUCUUCAAUAAUUGAACGUGUAGUCAUGAGGAAGCGAUAAAAUCGAAUUGCUUCCUUAAUGUGUGAAAUUG ---------...(((.((((....)).(((......)))...)).)))..(((((..(((..(.....((((((((((((........)))))))))))))..)))..))))) ( -24.70, z-score = -1.39, R) >droYak2.chr3R 12957349 103 - 28832112 ----------UCGGAAUGGGAACGCCGGCGGCAAAACGUAAACUCUUCAAUAAUUGAACGUGUAGUCAUGAGGAAGCGAUAAAAUCGAAUUGCUUCCUUAAUGUGUGAAAUUG ----------.(((...(....).)))(((......)))......((((...(((((.((((....)))).(((((((((........))))))))))))))...)))).... ( -25.60, z-score = -1.36, R) >droSec1.super_0 7794998 103 + 21120651 ----------UCGGAAAGGGAACGCCGCCGGCAAAACGUAAACUCUUCAAUAAUUGAACGUGUAGUCAUGAGGAAGCGAUAAAAUCGAAUUGCUUCCUUAAUGUGUGAAAUUG ----------((((...((.....)).))))..............((((...(((((.((((....)))).(((((((((........))))))))))))))...)))).... ( -25.10, z-score = -1.36, R) >droSim1.chr3R 14764083 103 + 27517382 ----------UCGGAAAGGGAACGCCGCCGGCAAAACGUAAACUCUUCAAUAAUUGAACGUGUAGUCAUGAGGAAGCGAUAAAAUCGAAUUGCUUCCUUAAUGUGUGAAAUUG ----------((((...((.....)).))))..............((((...(((((.((((....)))).(((((((((........))))))))))))))...)))).... ( -25.10, z-score = -1.36, R) >droVir3.scaffold_12855 5982020 102 + 10161210 ----------GGCAAAAGGAGGCGCAGACAGC-AAACGUAAACUCUUCAAUAAUUGAACGUGAAGUCAUGAGGAAGCGAUAAAAUCAAAUUGUUUCCUUAAUGUGUGAAAUUG ----------...........((((((((...-..((((.......(((.....)))))))...))).((((((((((((........)))))))))))).)))))....... ( -22.51, z-score = -1.29, R) >droMoj3.scaffold_6540 25625355 93 + 34148556 -------------------AGGCGCAGACAGC-AAACGUAAACUCUUCAAUAAUUGAACGUGAAGUCAUGAGGAAGCGAUAAAAUCAAAUUGCUUCCUUAAUGUGUGAAAUUG -------------------..((((((((...-..((((.......(((.....)))))))...))).((((((((((((........)))))))))))).)))))....... ( -24.91, z-score = -2.88, R) >droGri2.scaffold_14906 7392835 108 + 14172833 ----GGGAAACGGAGUCGAAGGCGUAGACAGC-AAACGUAAACUCUUCAAUAAUUGAACGUGAAGUCAUGAGGAAGCGAUAAAAUCAAAUUGCUUCCUUAAUGUGUGAAAUUG ----.(....)(((((((...((.......))-...))...)))))(((...(((((.((((....)))).(((((((((........))))))))))))))...)))..... ( -26.80, z-score = -2.08, R) >droWil1.scaffold_181089 11381137 99 - 12369635 ----------UCAA--CAUCAACG-CAACGGC-AAACGUAAACUCUUCAAUAAUUGAACGUGUAGUCAUGAGGAAGCGAUAAAAUCGAAUUGCUUCCUUAAUGUGUGAAAUUG ----------....--..(((.((-((..(((-..((((.......(((.....)))))))...))).((((((((((((........)))))))))))).)))))))..... ( -25.81, z-score = -3.06, R) >consensus __________UCGGAAAGGGAACGCCGACGGC_AAACGUAAACUCUUCAAUAAUUGAACGUGUAGUCAUGAGGAAGCGAUAAAAUCGAAUUGCUUCCUUAAUGUGUGAAAUUG .......................((.....)).............((((...(((((.((((....)))).(((((((((........))))))))))))))...)))).... (-19.69 = -19.72 + 0.02)
| Location | 8,663,750 – 8,663,853 |
|---|---|
| Length | 103 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 77.70 |
| Shannon entropy | 0.49146 |
| G+C content | 0.43132 |
| Mean single sequence MFE | -28.81 |
| Consensus MFE | -16.27 |
| Energy contribution | -16.20 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.984775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
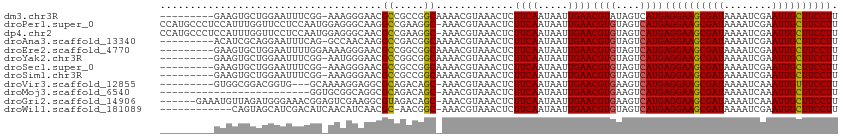
>dm3.chr3R 8663750 103 - 27905053 ---------GAAGUGCUGGAAUUUCGG-AAAGGGAACGCCGCCGGCAAAACGUAAACUCUUCAAUAAUUGAACGUAUAGUCAUGAGGAAGCGAUAAAAUCGAAUUGCUUCCUU ---------((((((((((.....(((-...(....).)))))))))...........)))).....................(((((((((((........))))))))))) ( -27.30, z-score = -1.61, R) >droPer1.super_0 2829939 112 + 11822988 CCAUGCCCUCCAUUUGGUUCCUCCAAUGGAGGGCAAGGCCGAAGGC-AAACGUAAACUCUUCAAUAAUUGAACGUGUAGUCAUGAGGAAGCGAUAAAAUCGAAUUGCUUCCUU ((.(((((((((((.((.....))))))))))))).)).....(((-..((((.......(((.....)))))))...)))..(((((((((((........))))))))))) ( -42.41, z-score = -4.80, R) >dp4.chr2 11525891 112 - 30794189 CCAUGCCCUCCAUUUGGUUCCUCCAAUGGAGGGCAACGCCGAAGGC-AAACGUAAACUCUUCAAUAAUUGAACGUGUAGUCAUGAGGAAGCGAUAAAAUCGAAUUGCUUCCUU ...(((((((((((.((.....)))))))))))))..(((...)))-............((((.....))))((((....))))((((((((((........)))))))))). ( -40.60, z-score = -4.69, R) >droAna3.scaffold_13340 14198721 103 + 23697760 ---------ACAUCGCAGGAAUUUCAG-GCCAACAAGGCCGACGGCAAAACGUAAACUCUUCAAUAAUUGAACGUGUAGUCAUGAGGAAGCGAUAAAAUCGAAUUGCUUCCUU ---------.....(((.(...(((((-(((.....)))).(((......)))...............))))).)))......(((((((((((........))))))))))) ( -27.60, z-score = -2.25, R) >droEre2.scaffold_4770 4761431 104 + 17746568 ---------GAAGUGCUGGAAUUUUGGAAAAGGGAACGCCGGCGGCAAAACGUAAACUCUUCAAUAAUUGAACGUGUAGUCAUGAGGAAGCGAUAAAAUCGAAUUGCUUCCUU ---------.....(((((..((((....)))).....)))))(((...((((.......(((.....)))))))...)))..(((((((((((........))))))))))) ( -27.71, z-score = -1.70, R) >droYak2.chr3R 12957363 103 - 28832112 ---------GAAGUGCUGGAAUUUCGG-AAUGGGAACGCCGGCGGCAAAACGUAAACUCUUCAAUAAUUGAACGUGUAGUCAUGAGGAAGCGAUAAAAUCGAAUUGCUUCCUU ---------.....(((((..((((..-....))))..)))))(((...((((.......(((.....)))))))...)))..(((((((((((........))))))))))) ( -30.01, z-score = -2.19, R) >droSec1.super_0 7795012 103 + 21120651 ---------GAAGUGCUGGAAUUUCGG-AAAGGGAACGCCGCCGGCAAAACGUAAACUCUUCAAUAAUUGAACGUGUAGUCAUGAGGAAGCGAUAAAAUCGAAUUGCUUCCUU ---------((((((((((.....(((-...(....).)))))))))...........))))..........((((....))))((((((((((........)))))))))). ( -28.70, z-score = -1.78, R) >droSim1.chr3R 14764097 103 + 27517382 ---------GAAGUGCUGGAAUUUCGG-AAAGGGAACGCCGCCGGCAAAACGUAAACUCUUCAAUAAUUGAACGUGUAGUCAUGAGGAAGCGAUAAAAUCGAAUUGCUUCCUU ---------((((((((((.....(((-...(....).)))))))))...........))))..........((((....))))((((((((((........)))))))))). ( -28.70, z-score = -1.78, R) >droVir3.scaffold_12855 5982034 100 + 10161210 ---------GUGGCGGACGGUG---GCAAAAGGAGGCGCAGACAGC-AAACGUAAACUCUUCAAUAAUUGAACGUGAAGUCAUGAGGAAGCGAUAAAAUCAAAUUGUUUCCUU ---------(((((..((((((---.(.......).))).......-............((((.....)))))))...)))))(((((((((((........))))))))))) ( -22.60, z-score = -0.46, R) >droMoj3.scaffold_6540 25625369 87 + 34148556 -------------------------GGUGCGGCAGGCGCAGACAGC-AAACGUAAACUCUUCAAUAAUUGAACGUGAAGUCAUGAGGAAGCGAUAAAAUCAAAUUGCUUCCUU -------------------------.((((.....)))).(((...-..((((.......(((.....)))))))...)))..(((((((((((........))))))))))) ( -22.91, z-score = -1.59, R) >droGri2.scaffold_14906 7392849 106 + 14172833 ------GAAAUGUUAGAUGGGAAACGGAGUCGAAGGCGUAGACAGC-AAACGUAAACUCUUCAAUAAUUGAACGUGAAGUCAUGAGGAAGCGAUAAAAUCAAAUUGCUUCCUU ------((.(((((.(((.(....)...)))(((((.((..((...-....))..)))))))........)))))....))..(((((((((((........))))))))))) ( -26.10, z-score = -2.03, R) >droWil1.scaffold_181089 11381151 99 - 12369635 ------------CAGUAGCAUCGACAUCAACAUCAACGC-AACGGC-AAACGUAAACUCUUCAAUAAUUGAACGUGUAGUCAUGAGGAAGCGAUAAAAUCGAAUUGCUUCCUU ------------..........(((..............-.(((..-...)))..((.(((((.....)))).).)).)))..(((((((((((........))))))))))) ( -21.10, z-score = -1.43, R) >consensus _________GAAGUGCUGGAAUUUCGG_AAAGGGAACGCCGACGGC_AAACGUAAACUCUUCAAUAAUUGAACGUGUAGUCAUGAGGAAGCGAUAAAAUCGAAUUGCUUCCUU .....................................((.....)).............((((.....))))((((....))))((((((((((........)))))))))). (-16.27 = -16.20 + -0.07)
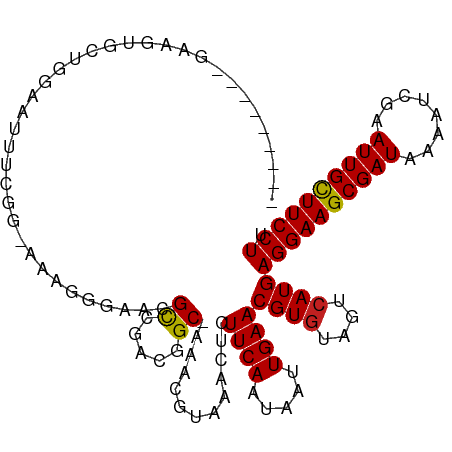
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:10:22 2011