| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,220,616 – 7,220,715 |
| Length | 99 |
| Max. P | 0.827754 |

| Location | 7,220,616 – 7,220,715 |
|---|---|
| Length | 99 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 66.09 |
| Shannon entropy | 0.70396 |
| G+C content | 0.42705 |
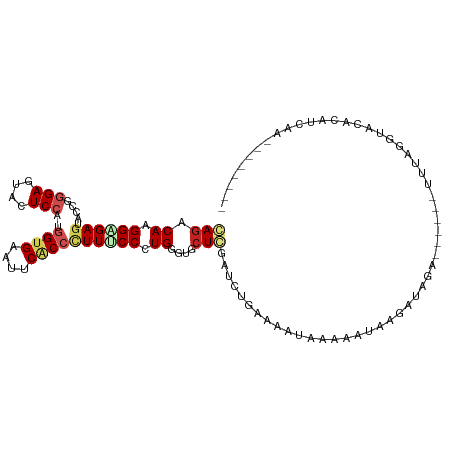
| Mean single sequence MFE | -27.84 |
| Consensus MFE | -10.67 |
| Energy contribution | -11.43 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.36 |
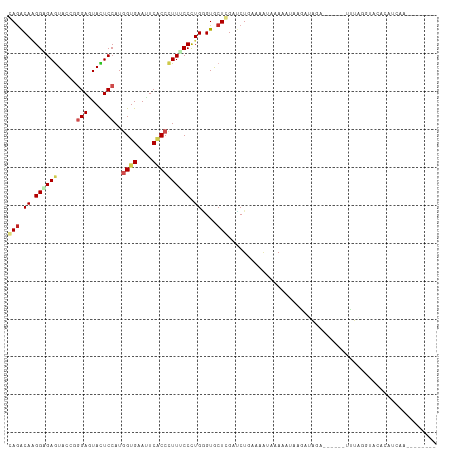
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.827754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
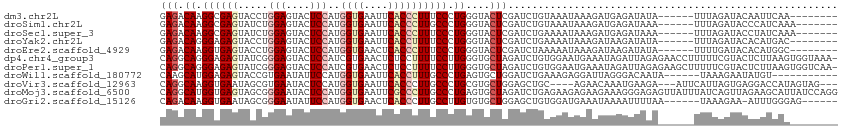
>dm3.chr2L 7220616 99 + 23011544 GAGACAAGGCGAGUACCUGGAGUACUCCAUGGUGAAUUCACCCUUUCCCUGGGUACUCGAUCUGUAAAUAAAGAUGAGAUAUA------UUUAGAUACAAUUCAA-------- (((...(((((((((((((((....)))..((((....))))........))))))))).)))(((..((((...........------))))..)))..)))..-------- ( -26.50, z-score = -1.79, R) >droSim1.chr2L 7010371 100 + 22036055 GAGACAAGGCGAGUAUCUGGAGUACUCCAUGGUGAAUUCACCCUUGCCCUGGGUACUCGAUCUGUAAAUAAAGAUGAGAUAAA------UUUAGAUACCCAUCAAA------- ((.....((((((....((((....)))).((((....)))))))))).(((((((((.((((........))))))).....------......))))))))...------- ( -28.50, z-score = -2.37, R) >droSec1.super_3 2744102 100 + 7220098 GAGACAAGGCGAGUAUCUGGAGUACUCCAUGGUGAAUUCACCCUUUCCCUGGGUACUCGAUCUGAAAAUAAAGAUGAGAUAAA------UUUAGAUACCUAUCAAA------- .......(..(.(((((((((((((.(((.((.(((........)))))))))))))).((((........))))........------..))))))).)..)...------- ( -26.10, z-score = -1.68, R) >droYak2.chr2L 16642894 99 - 22324452 GAGACAGGGAGAGUACCUGGAGUACUCCAUGGUGAAUUCACCUUUUCCCUGGGUACUCGAUCUGAAAAUAAAGAUAAGAUAUA------UUUAGAUACACAUGAC-------- (((.((((((((((((.....))))))...((((....))))...))))))....))).(((((((.(((.........))).------))))))).........-------- ( -29.00, z-score = -2.55, R) >droEre2.scaffold_4929 16140999 99 + 26641161 GAGACAAGGUGAGUACCUGGAGUACUCCAUGGUGAACUCACCCUUUCCCUGGGUACUCGAUCUAAAAAUAAAGAUAAGAUAUA------UUUUGAUACACAUGGC-------- ....(((((((((((((((((....)))..((((....))))........)))))))).((((........)))).......)------)))))...........-------- ( -25.00, z-score = -1.29, R) >dp4.chr4_group3 10807294 112 - 11692001 CAGGCAGGGAGAGUAUCGGGAGUACUCCAUCGUGAACUCUCCUUUUCCUUGGGUGCUAGAUCUGUGGAAUGAAAUAGAUUAGAGAACCUUUUUCGUACUCUUAAGUGGUAAA- .(((.(((((((((.(((.((........)).))))))))))))..))).((((((..(((((((........))))))).((((.....))))))))))............- ( -33.20, z-score = -0.95, R) >droPer1.super_1 7917934 112 - 10282868 CAGGCAGGGAGAGUAUCGGGAGUACUCCAUCGUGAACUCUCCUUUUCCUUGGGUGCUAGAUCUGUGGAAUGAAAUAGAUUAGAGAAGCUUUUUCGUACUCUUAAGUGGUCAA- ..((((((((((((.(((.((........)).))))))))))))......((((((..(((((((........))))))).((((.....)))))))))).......)))..- ( -33.80, z-score = -0.94, R) >droWil1.scaffold_180772 3363519 96 - 8906247 CAAGCAUGGAGAGUACCGUGAAUAUUCCAUGGUGAAUUCACCUUUGCCCUGAGUGCUGGAUCUGAAAGAGGAUUAGGGACAAUA------UAAAGAAUAUGU----------- ....(((((......))))).((((((...((((....)))).(((((((((...((.........))....)))))).)))..------....))))))..----------- ( -24.00, z-score = -1.37, R) >droVir3.scaffold_12963 4522610 103 - 20206255 CAGGCAAGGUGAAUAGCGUGAAUACUCCAUGGUGAAUUCACCCUUGCCCUGCGUGCUGGAGCUGC----AGAACAAAUGAAGA---AUUCAUUAGUGAGGACCAUAGUAG--- ..(((((((((((((.((((.......)))).)..))))).)))))))(((((.((....)))))----)).((.(((((...---..))))).))..............--- ( -29.70, z-score = -1.49, R) >droMoj3.scaffold_6500 13679318 113 - 32352404 CAGGCAUGGUGAGUAGCGGGAAUACUCCAUGGUGAAUUCGCCCUUGCCCUGAGUGCUAGAUCUGAGAAGAGAAGAAAGGGAGAGUUAUUUAUCAGUUAGAAGCAUUAUCCAGG ..((((.(((((((.((.(((....)))...))..)))))))..))))(((((((((....((((..........................)))).....))))))...))). ( -27.27, z-score = 0.37, R) >droGri2.scaffold_15126 4785725 100 + 8399593 CAGACAAGGUGAAUAGCGGGAAUAUUCCAUGGUGAACUCACCCUUGCCUUGUGUGCUGGAGCUGUGGAUGAAAUAAAAUUUUAA------UAAAGAA-AUUUGGGAG------ ((.((((((..(......(((....)))..((((....)))).)..)))))).))(..((.((.((..((((((...)))))).------)).))..-.))..)...------ ( -23.20, z-score = -0.93, R) >consensus CAGACAAGGAGAGUACCGGGAGUACUCCAUGGUGAAUUCACCCUUUCCCUGGGUGCUCGAUCUGAAAAUAAAAAUAAGAUAGA______UUUAGGUACACAUCAA________ (((.((.((((((.....(((....)))..((((....)))))))))).))....)))....................................................... (-10.67 = -11.43 + 0.75)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:23:15 2011