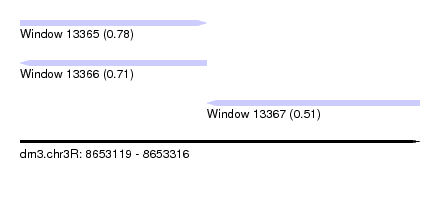
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,653,119 – 8,653,316 |
| Length | 197 |
| Max. P | 0.781414 |

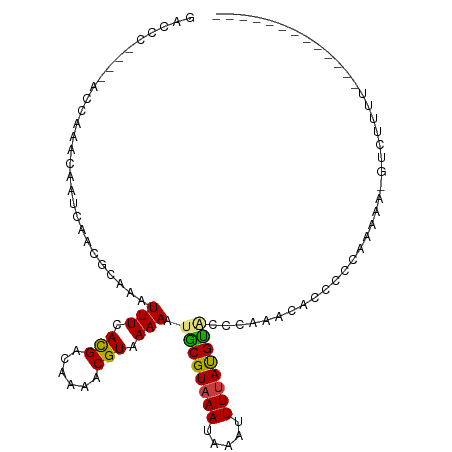
| Location | 8,653,119 – 8,653,211 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 73.34 |
| Shannon entropy | 0.47753 |
| G+C content | 0.38041 |
| Mean single sequence MFE | -20.77 |
| Consensus MFE | -11.97 |
| Energy contribution | -12.06 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.781414 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8653119 92 + 27905053 -------------AAAAUACAUUUUUGGGGGUGUUUGGGUGCAUAAAAUUUAUUUACGCAUUUUUACGUUUUGCCGUGAAAUUUGCGUUGAUUGUUUGGU----GGGUC -------------.((((((..........)))))).....(((.((((..(((.(((((.(((((((......)))))))..))))).))).)))).))----).... ( -18.00, z-score = -0.47, R) >droAna3.scaffold_13340 14188391 105 - 23697760 AAAUGUUUCGUUUUUUUUGUUUUUGUUGGGGUGUUGGAGCACAUAAAAUUUAUUUACGCAUUUUUACGUUUUGUCGUGAAAUUUGCGUUGAUUGUUUGGA----GGGUC ..((.((((.....................((((....))))...((((..(((.(((((.(((((((......)))))))..))))).))).)))))))----).)). ( -19.60, z-score = -0.87, R) >droEre2.scaffold_4770 4750405 92 - 17746568 -------------AACAGACAUUUUUUGGGGUGUUUGGGUGCAUAAAAUUUAUUUACGCAUUUUUACGUUUUGUCGUGAAAUUUGCGUUGAUUGUUUGGU----GGGUC -------------..(((((((((....)))))))))....(((.((((..(((.(((((.(((((((......)))))))..))))).))).)))).))----).... ( -24.20, z-score = -3.24, R) >droSim1.chr3R 14753237 91 - 27517382 -------------AAAAGAC-UUUUUGGCGGUGUUUGGGUGCAUAAAAUUUAUUUACGCAUUUUUACGUUUUGUCGUGAAAUUUGCGUUGAUUGUUUGGU----GGGUC -------------....(((-(..(..(((((((......)))).......(((.(((((.(((((((......)))))))..))))).))))).)..).----.)))) ( -19.80, z-score = -1.38, R) >dp4.chr2 19259094 88 - 30794189 ---------------------UCGGGGAAAGUGUUGGGGGACAUAAAAUUUAUUUACGUAUUUUUACGUUGCGUCGUGAAAUUUGCGUUGAUUGUUUGGUCGCUGGGUC ---------------------............((.((.(((...((((..(((.(((((.(((((((......)))))))..))))).))).)))).))).)).)).. ( -19.80, z-score = -1.04, R) >droPer1.super_0 8983843 88 + 11822988 ---------------------UCGGGGAAAGUGUUGGGGGACAUAAAAUUUAUUUACGUAUUUUUACGUUGCGUCGUGAAAUUUGCGUUGAUUGUUUGGUCGCUGGGUC ---------------------............((.((.(((...((((..(((.(((((.(((((((......)))))))..))))).))).)))).))).)).)).. ( -19.80, z-score = -1.04, R) >droGri2.scaffold_14906 7383386 93 - 14172833 -------------AAAGGGCAACUCUUCGGCCAAAUCGUUACGUCAAAUGUAUUUACGCAGUUUUACGUUUUGUCAUGAAAUGUGCGUUGAUUGUUAGCC--CUUGCU- -------------.(((((((((...(((((((..((((.(((..(((((((............)))))))))).))))....)).)))))..))).)))--)))...- ( -24.20, z-score = -1.64, R) >consensus _____________AAAAGAC_UUUGUGGGGGUGUUUGGGUACAUAAAAUUUAUUUACGCAUUUUUACGUUUUGUCGUGAAAUUUGCGUUGAUUGUUUGGU____GGGUC ..............................((((......)))).((((..(((.(((((.(((((((......)))))))..))))).))).))))............ (-11.97 = -12.06 + 0.08)

| Location | 8,653,119 – 8,653,211 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 73.34 |
| Shannon entropy | 0.47753 |
| G+C content | 0.38041 |
| Mean single sequence MFE | -10.48 |
| Consensus MFE | -6.46 |
| Energy contribution | -6.11 |
| Covariance contribution | -0.34 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.710544 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8653119 92 - 27905053 GACCC----ACCAAACAAUCAACGCAAAUUUCACGGCAAAACGUAAAAAUGCGUAAAUAAAUUUUAUGCACCCAAACACCCCCAAAAAUGUAUUUU------------- .....----..............(((..(((.(((......))).))).((((((((.....))))))))..................))).....------------- ( -10.60, z-score = -1.18, R) >droAna3.scaffold_13340 14188391 105 + 23697760 GACCC----UCCAAACAAUCAACGCAAAUUUCACGACAAAACGUAAAAAUGCGUAAAUAAAUUUUAUGUGCUCCAACACCCCAACAAAAACAAAAAAAACGAAACAUUU .....----.........((.(((((..(((.(((......))))))..))))).............(((......))).....................))....... ( -8.60, z-score = -1.05, R) >droEre2.scaffold_4770 4750405 92 + 17746568 GACCC----ACCAAACAAUCAACGCAAAUUUCACGACAAAACGUAAAAAUGCGUAAAUAAAUUUUAUGCACCCAAACACCCCAAAAAAUGUCUGUU------------- .....----....((((..((.......(((.(((......))).))).((((((((.....))))))))..................))..))))------------- ( -10.30, z-score = -1.65, R) >droSim1.chr3R 14753237 91 + 27517382 GACCC----ACCAAACAAUCAACGCAAAUUUCACGACAAAACGUAAAAAUGCGUAAAUAAAUUUUAUGCACCCAAACACCGCCAAAAA-GUCUUUU------------- (((..----...................(((.(((......))).))).((((((((.....))))))))..................-)))....------------- ( -10.40, z-score = -2.00, R) >dp4.chr2 19259094 88 + 30794189 GACCCAGCGACCAAACAAUCAACGCAAAUUUCACGACGCAACGUAAAAAUACGUAAAUAAAUUUUAUGUCCCCCAACACUUUCCCCGA--------------------- (((...(((.............)))...............(((((....))))).............)))..................--------------------- ( -6.92, z-score = -0.55, R) >droPer1.super_0 8983843 88 - 11822988 GACCCAGCGACCAAACAAUCAACGCAAAUUUCACGACGCAACGUAAAAAUACGUAAAUAAAUUUUAUGUCCCCCAACACUUUCCCCGA--------------------- (((...(((.............)))...............(((((....))))).............)))..................--------------------- ( -6.92, z-score = -0.55, R) >droGri2.scaffold_14906 7383386 93 + 14172833 -AGCAAG--GGCUAACAAUCAACGCACAUUUCAUGACAAAACGUAAAACUGCGUAAAUACAUUUGACGUAACGAUUUGGCCGAAGAGUUGCCCUUU------------- -...(((--(((.(((..((..((..((..((........(((((....)))))...(((.......)))..))..))..))..))))))))))).------------- ( -19.60, z-score = -0.96, R) >consensus GACCC____ACCAAACAAUCAACGCAAAUUUCACGACAAAACGUAAAAAUGCGUAAAUAAAUUUUAUGUACCCAAACACCCCCAAAAA_GUCUUUU_____________ ............................(((.(((......))).))).((((((((.....))))))))....................................... ( -6.46 = -6.11 + -0.34)
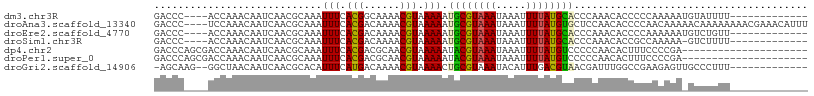

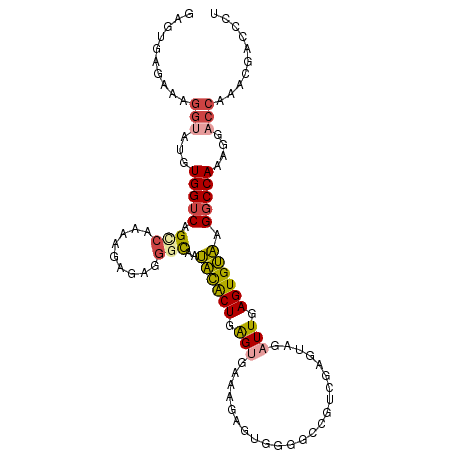
| Location | 8,653,211 – 8,653,316 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 77.76 |
| Shannon entropy | 0.43646 |
| G+C content | 0.51858 |
| Mean single sequence MFE | -26.50 |
| Consensus MFE | -13.57 |
| Energy contribution | -13.21 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.513823 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8653211 105 - 27905053 GAGUGAGAAAGGUAUGUGGUCAGCCGAAAGAGAGAGCAAUACGCUGAGUGAAAGAGUGGGGCCGACGAGUAGAUUGAGUGUAAGGCCAAAGGACCAAACGACCCU ..........(((...(((((...(....)...(.((..((((((.(((......(((....).))......))).))))))..)))....)))))....))).. ( -23.90, z-score = -1.25, R) >droEre2.scaffold_4770 4750497 104 + 17746568 GAAUGAGAAAGGUAUGUGGUCAGACAAAAGAGACGGCAAUACACUGAGUGGAAGAGUGGAGCCGUCGAGUAGAUUGAGUGUAAGGCCAAAGUACC-AACGACCCU ..........(((((.(((((..(((.((..((((((..(((.((.......)).)))..)))))).......))...)))..)))))..)))))-......... ( -28.50, z-score = -2.96, R) >droYak2.chr3R 12946312 105 - 28832112 GAAUGAGAAAGGUAUGUGGUCAGCCAAACGAGAGGGCAAUAUACUGAGGGGCAGAGUGGAGCCGUCGAGUAGAUUGAGUGUAAGGCCAAAGGACCAAAAGACCCU ..........(((...(((((.......(....)(((..((((((.(..(((........)))(((.....)))).))))))..)))....)))))....))).. ( -28.40, z-score = -1.96, R) >droSec1.super_0 7784435 105 + 21120651 GAGUGAGAAAGGUAUGUGGUCAGUCAAAAGAGAGGGCAAUACACUGAGUGAAAGAGUGGGGCCGUCGAGUAGAUUGAGUGUAAGGCCAAAGGACCAAACGACCCU ..........(((...(((((..((......)).(((..((((((.(((.(..((.((....))))...)..))).))))))..)))....)))))....))).. ( -26.00, z-score = -1.43, R) >droSim1.chr3R 14753328 105 + 27517382 GAGUGAGAAAGGAAUGUGGUCAGCCGAAAGAGAGGGCAAUACACUGAGUGAAAGAGUGGGGCAGUCGAGUAGAUUGAGUGUAGGGCCAAAGGACCCAACGACCCU .........(((..(((((((...(....)....(((..((((((.(((.(..((.((...)).))...)..))).))))))..)))....))))..)))..))) ( -26.70, z-score = -1.30, R) >droPer1.super_0 8983931 81 - 11822988 -------GAAGAGAGGUGG-CAGCCGA---GGAGGCUGCCUUACUGGGCAA----------UUCUGGAGGAGAUUGAGUGAGAGGCCAAGGAGC---AGGACCCU -------....((.(((((-(((((..---...)))))))...((.(((..----------..((.((.....)).))......))).))....---...))))) ( -25.50, z-score = -0.46, R) >consensus GAGUGAGAAAGGUAUGUGGUCAGCCAAAAGAGAGGGCAAUACACUGAGUGAAAGAGUGGGGCCGUCGAGUAGAUUGAGUGUAAGGCCAAAGGACCAAACGACCCU .................((((.............(((..((((((.(((.......................))).))))))..)))............)))).. (-13.57 = -13.21 + -0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:10:16 2011