
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,646,476 – 8,646,596 |
| Length | 120 |
| Max. P | 0.652527 |

| Location | 8,646,476 – 8,646,594 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 88.01 |
| Shannon entropy | 0.21747 |
| G+C content | 0.31700 |
| Mean single sequence MFE | -25.72 |
| Consensus MFE | -16.82 |
| Energy contribution | -19.40 |
| Covariance contribution | 2.58 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.540076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8646476 118 + 27905053 UAAACAGUGGUGUGCACUUAUUUUUGAUAAUUUACACAUUUUUAUUAUCUGGAAACUAAUAAAUAAGUGUCUGAACG-UGUAAUCGAUAAGAGCACAUUUUUCACCCAGUGCACUUUAU .....((((((((((.((((((....(((.((((.((((((...((((..(....)..))))..)))))).))))..-)))....)))))).))))))....(((...))))))).... ( -24.60, z-score = -1.64, R) >droSim1.chr3R 14746590 118 - 27517382 UAAAGAGUUGUGUGCACUUAUUUCUGAUAAUUUACACAUGUUUAUUAUCUGGAAACUAAUAAAUAUGUGUCUGAACC-UAUAAUCGAUAAGAACACAUUUUUCACCCAGUGCACUUUAU ....(((..(((((..((((((....(((.(((((((((((((((((...(....)))))))))))))))..)))..-)))....))))))..)))))..)))................ ( -27.00, z-score = -3.18, R) >droSec1.super_0 7777874 119 - 21120651 UAAAGAGUAGUGUGCACUUAUUUCUGAUAAUUUACACAUGUUUAUUAUCUGGAAACUAAUAAAUAUGUGUCUGAACCGUGUAAUCGAUAAGAACACAUUUUUCACCCAGUGCACUUUAU ......((((.(((((((...............((((((((((((((...(....))))))))))))))).((((..((((..((.....))..))))..))))...))))))).)))) ( -30.70, z-score = -3.63, R) >droYak2.chr3R 12939213 118 + 28832112 UGUAUAGAAGGGAGAACUCAUUUGUGAUAACUUACACAUAUUUAUUAUCUGUGAACCAAUAAAUAUGUGUCUGAACC-UGAAAUCGAUAAGAACACAUUUUUCACCCAGUGCACUGUAU (((((....(((.(((......((((....(..(((((((((((((....(....).)))))))))))))..)....-.....((.....)).))))...))).))).)))))...... ( -22.60, z-score = -0.66, R) >droEre2.scaffold_4770 4743575 117 - 17746568 UUUACAGGAGUGUGCACUUGUUUGUGAUAAUUUACACAUGUUUAUUAUCAGUAAACUAAUAAAUAAGCCUCUGAACC-UGUAAUCGAUAAGAACCCAUUUU-CACCCAGUGCACUUUGU ......((((((..(.(((((((((((((((..((....))..))))))((....)).)))))))))....((((..-((...((.....))...))..))-))....)..)))))).. ( -23.70, z-score = -1.46, R) >consensus UAAACAGUAGUGUGCACUUAUUUCUGAUAAUUUACACAUGUUUAUUAUCUGGAAACUAAUAAAUAUGUGUCUGAACC_UGUAAUCGAUAAGAACACAUUUUUCACCCAGUGCACUUUAU ...........(((((((...............((((((((((((((...(....))))))))))))))).((((...(((..((.....))..)))...))))...)))))))..... (-16.82 = -19.40 + 2.58)

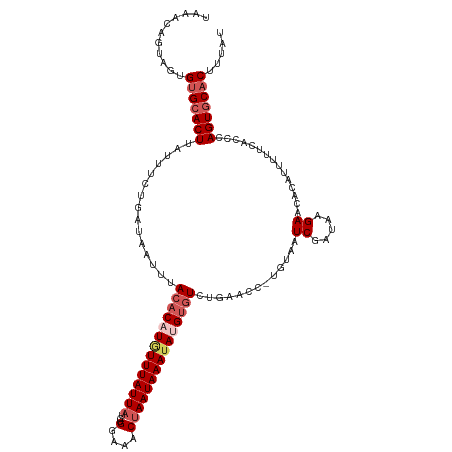
| Location | 8,646,476 – 8,646,594 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 88.01 |
| Shannon entropy | 0.21747 |
| G+C content | 0.31700 |
| Mean single sequence MFE | -26.49 |
| Consensus MFE | -19.82 |
| Energy contribution | -20.94 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.652527 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8646476 118 - 27905053 AUAAAGUGCACUGGGUGAAAAAUGUGCUCUUAUCGAUUACA-CGUUCAGACACUUAUUUAUUAGUUUCCAGAUAAUAAAAAUGUGUAAAUUAUCAAAAAUAAGUGCACACCACUGUUUA .....(((((((...((((...((((.((.....)).))))-..)))).((((...(((((((.........)))))))...))))...............)))))))........... ( -23.20, z-score = -1.26, R) >droSim1.chr3R 14746590 118 + 27517382 AUAAAGUGCACUGGGUGAAAAAUGUGUUCUUAUCGAUUAUA-GGUUCAGACACAUAUUUAUUAGUUUCCAGAUAAUAAACAUGUGUAAAUUAUCAGAAAUAAGUGCACACAACUCUUUA .....(((((((.((((((..(((((((...(((.......-)))...))))))).)))))).(((((..((((((..((....))..)))))).))))).)))))))........... ( -27.60, z-score = -2.63, R) >droSec1.super_0 7777874 119 + 21120651 AUAAAGUGCACUGGGUGAAAAAUGUGUUCUUAUCGAUUACACGGUUCAGACACAUAUUUAUUAGUUUCCAGAUAAUAAACAUGUGUAAAUUAUCAGAAAUAAGUGCACACUACUCUUUA .....(((((((.((((((..(((((((...((((......))))...))))))).)))))).(((((..((((((..((....))..)))))).))))).)))))))........... ( -29.40, z-score = -2.90, R) >droYak2.chr3R 12939213 118 - 28832112 AUACAGUGCACUGGGUGAAAAAUGUGUUCUUAUCGAUUUCA-GGUUCAGACACAUAUUUAUUGGUUCACAGAUAAUAAAUAUGUGUAAGUUAUCACAAAUGAGUUCUCCCUUCUAUACA .....(((....(((.(((...((((..(((...((.....-...))..((((((((((((((.........)))))))))))))))))....))))......))).))).....))). ( -25.00, z-score = -0.83, R) >droEre2.scaffold_4770 4743575 117 + 17746568 ACAAAGUGCACUGGGUG-AAAAUGGGUUCUUAUCGAUUACA-GGUUCAGAGGCUUAUUUAUUAGUUUACUGAUAAUAAACAUGUGUAAAUUAUCACAAACAAGUGCACACUCCUGUAAA .....((((((((.(((-((((((((((((.(((.......-)))..))).))))))))..((((((((...............))))))))))))...).)))))))........... ( -27.26, z-score = -1.52, R) >consensus AUAAAGUGCACUGGGUGAAAAAUGUGUUCUUAUCGAUUACA_GGUUCAGACACAUAUUUAUUAGUUUCCAGAUAAUAAACAUGUGUAAAUUAUCACAAAUAAGUGCACACUACUCUUUA .....(((((((...((((...((((.((.....)).))))...)))).((((((.(((((((.........))))))).))))))...............)))))))........... (-19.82 = -20.94 + 1.12)

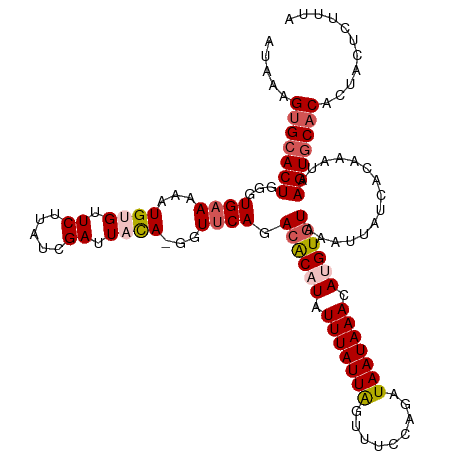

| Location | 8,646,478 – 8,646,596 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 88.60 |
| Shannon entropy | 0.20595 |
| G+C content | 0.32378 |
| Mean single sequence MFE | -25.98 |
| Consensus MFE | -16.82 |
| Energy contribution | -19.40 |
| Covariance contribution | 2.58 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.538432 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8646478 118 + 27905053 AACAGUGGUGUGCACUUAUUUUUGAUAAUUUACACAUUUUUAUUAUCUGGAAACUAAUAAAUAAGUGUCUGAACG-UGUAAUCGAUAAGAGCACAUUUUUCACCCAGUGCACUUUAUAG ...((((((((((.((((((....(((.((((.((((((...((((..(....)..))))..)))))).))))..-)))....)))))).))))))....(((...)))))))...... ( -24.60, z-score = -1.55, R) >droSim1.chr3R 14746592 118 - 27517382 AAGAGUUGUGUGCACUUAUUUCUGAUAAUUUACACAUGUUUAUUAUCUGGAAACUAAUAAAUAUGUGUCUGAACC-UAUAAUCGAUAAGAACACAUUUUUCACCCAGUGCACUUUAUAG ..(((..(((((..((((((....(((.(((((((((((((((((...(....)))))))))))))))..)))..-)))....))))))..)))))..))).................. ( -27.00, z-score = -2.93, R) >droSec1.super_0 7777876 119 - 21120651 AAGAGUAGUGUGCACUUAUUUCUGAUAAUUUACACAUGUUUAUUAUCUGGAAACUAAUAAAUAUGUGUCUGAACCGUGUAAUCGAUAAGAACACAUUUUUCACCCAGUGCACUUUAUAG ....((((.(((((((...............((((((((((((((...(....))))))))))))))).((((..((((..((.....))..))))..))))...))))))).)))).. ( -31.40, z-score = -3.70, R) >droYak2.chr3R 12939215 118 + 28832112 UAUAGAAGGGAGAACUCAUUUGUGAUAACUUACACAUAUUUAUUAUCUGUGAACCAAUAAAUAUGUGUCUGAACC-UGAAAUCGAUAAGAACACAUUUUUCACCCAGUGCACUGUAUAG .......(((.(((......((((....(..(((((((((((((....(....).)))))))))))))..)....-.....((.....)).))))...))).))).............. ( -21.50, z-score = -0.68, R) >droEre2.scaffold_4770 4743577 117 - 17746568 UACAGGAGUGUGCACUUGUUUGUGAUAAUUUACACAUGUUUAUUAUCAGUAAACUAAUAAAUAAGCCUCUGAACC-UGUAAUCGAUAAGAACCCAUUUU-CACCCAGUGCACUUUGUAG (((((.((((..(.(((((((((((((((..((....))..))))))((....)).)))))))))....((((..-((...((.....))...))..))-))....)..))))))))). ( -25.40, z-score = -1.95, R) >consensus AACAGUAGUGUGCACUUAUUUCUGAUAAUUUACACAUGUUUAUUAUCUGGAAACUAAUAAAUAUGUGUCUGAACC_UGUAAUCGAUAAGAACACAUUUUUCACCCAGUGCACUUUAUAG .........(((((((...............((((((((((((((...(....))))))))))))))).((((...(((..((.....))..)))...))))...)))))))....... (-16.82 = -19.40 + 2.58)



| Location | 8,646,478 – 8,646,596 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 88.60 |
| Shannon entropy | 0.20595 |
| G+C content | 0.32378 |
| Mean single sequence MFE | -26.29 |
| Consensus MFE | -19.82 |
| Energy contribution | -20.94 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.541968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8646478 118 - 27905053 CUAUAAAGUGCACUGGGUGAAAAAUGUGCUCUUAUCGAUUACA-CGUUCAGACACUUAUUUAUUAGUUUCCAGAUAAUAAAAAUGUGUAAAUUAUCAAAAAUAAGUGCACACCACUGUU .......(((((((...((((...((((.((.....)).))))-..)))).((((...(((((((.........)))))))...))))...............)))))))......... ( -23.20, z-score = -1.36, R) >droSim1.chr3R 14746592 118 + 27517382 CUAUAAAGUGCACUGGGUGAAAAAUGUGUUCUUAUCGAUUAUA-GGUUCAGACACAUAUUUAUUAGUUUCCAGAUAAUAAACAUGUGUAAAUUAUCAGAAAUAAGUGCACACAACUCUU .......(((((((.((((((..(((((((...(((.......-)))...))))))).)))))).(((((..((((((..((....))..)))))).))))).)))))))......... ( -27.60, z-score = -2.54, R) >droSec1.super_0 7777876 119 + 21120651 CUAUAAAGUGCACUGGGUGAAAAAUGUGUUCUUAUCGAUUACACGGUUCAGACACAUAUUUAUUAGUUUCCAGAUAAUAAACAUGUGUAAAUUAUCAGAAAUAAGUGCACACUACUCUU .......(((((((.((((((..(((((((...((((......))))...))))))).)))))).(((((..((((((..((....))..)))))).))))).)))))))......... ( -29.40, z-score = -2.93, R) >droYak2.chr3R 12939215 118 - 28832112 CUAUACAGUGCACUGGGUGAAAAAUGUGUUCUUAUCGAUUUCA-GGUUCAGACACAUAUUUAUUGGUUCACAGAUAAUAAAUAUGUGUAAGUUAUCACAAAUGAGUUCUCCCUUCUAUA ..............(((.(((...((((..(((...((.....-...))..((((((((((((((.........)))))))))))))))))....))))......))).)))....... ( -24.00, z-score = -0.58, R) >droEre2.scaffold_4770 4743577 117 + 17746568 CUACAAAGUGCACUGGGUG-AAAAUGGGUUCUUAUCGAUUACA-GGUUCAGAGGCUUAUUUAUUAGUUUACUGAUAAUAAACAUGUGUAAAUUAUCACAAACAAGUGCACACUCCUGUA .......((((((((.(((-((((((((((((.(((.......-)))..))).))))))))..((((((((...............))))))))))))...).)))))))......... ( -27.26, z-score = -1.35, R) >consensus CUAUAAAGUGCACUGGGUGAAAAAUGUGUUCUUAUCGAUUACA_GGUUCAGACACAUAUUUAUUAGUUUCCAGAUAAUAAACAUGUGUAAAUUAUCACAAAUAAGUGCACACUACUCUU .......(((((((...((((...((((.((.....)).))))...)))).((((((.(((((((.........))))))).))))))...............)))))))......... (-19.82 = -20.94 + 1.12)

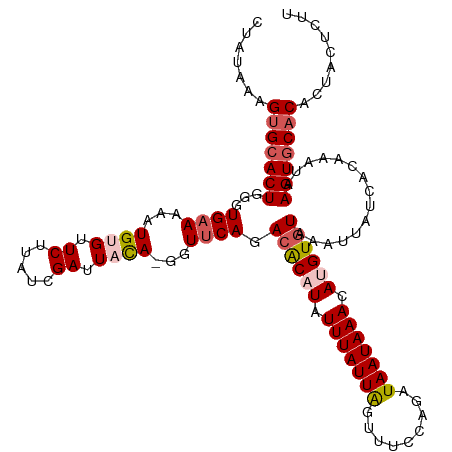
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:10:12 2011